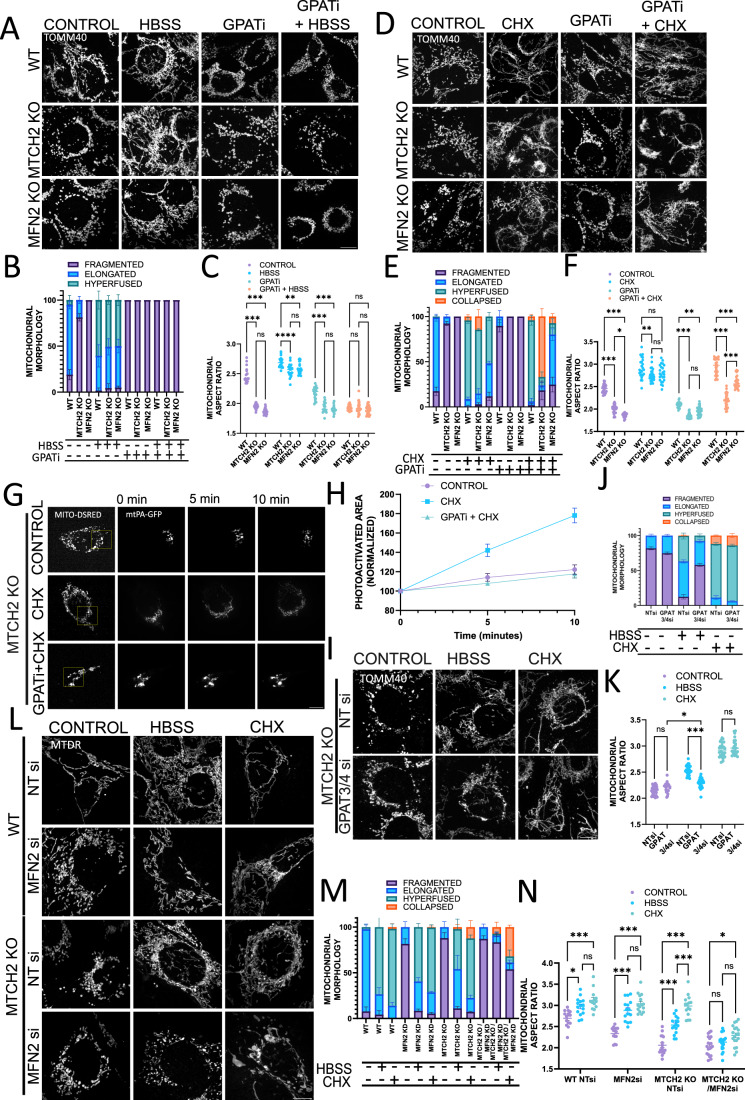
Figure 4. MTCH2 cooperates with MFN2 and LPA synthesis to sustain mitochondrial fusion under cellular stress conditions.
(A) Representatives images of WT, MTCH2 KO, and MFN2 KO MEFs either non-treated (control), HBSS treated for 4 h, treated with GPATi for 16 h, or pretreated for 16 h with GPATi and then treated with HBSS for 4 h in the presence of the inhibitor. Mitochondria were stained with αTOMM40 antibody. Scale bar 10 µm. (B) Classical classification of mitochondrial network morphology. Results are presented as average and SEM of three separate experiments; at least 20 cells were analyzed per condition. (C) Mitochondrial aspect ratio analysis. At least 25 fields were analyzed per condition. One-way ANOVA statistical analysis and SEM are shown. (D) Representatives images of WT, MTCH2 KO, and MFN2 KO MEFs either non-treated (control), treated with 10 μM CHX for 4 h, treated with GPATi for 16 h, or pretreated for 16 h with GPATi and then treated with 10 μM CHX for 4 h in the presence of the inhibitor. Mitochondria were stained with αTOMM40 antibody. Scale bar 10 µm. (E) Classical classification of mitochondrial network morphology. Results are presented as average and SEM of three separate experiments; at least 20 cells were analyzed per condition. (F) Mitochondrial aspect ratio analysis. At least 25 fields were analyzed per condition. One-way ANOVA statistical analysis and SEM are shown. (G) Mitochondrial fusion assay. MTCH2 KO MEFs were co-transfected with mt-PA-GFP and mito-dsRed (both constructs are targeted to the mitochondrial matrix). After transfection cells were either left untreated, treated with 10 μM CHX for 4 h, or pretreated for 16 h with GPATi and then treated with 10 μM CHX for 4 h in the presence of the inhibitor. Yellow box indicates the initially photoactivated area. After photoactivation, the GFP signal was followed for 10 min. (H) Quantification of mitochondrial fusion. Eight cells were analyzed in each condition, the GFP fluorescent area was calculated on time 0, 5, and 10 min after photoactivation, and the area was then normalized to the initially photoactivated area. Each data point shows average and SEM. (I) Representatives images of MTCH2 KO MEFs treated with non-targeting (NT) siRNA or a combination of GPAT3 and GPAT4 siRNAs, control or treated with HBSS for 4 h, or treated with 10 μM CHX for 4 h. Mitochondria were stained with αTOMM40 antibody. Scale bar 10 µm. (J) Classical classification of mitochondrial network morphology. Results are presented as average and SEM of three different experiments, and at least 30 cells were analyzed per condition. (K) Mitochondrial aspect ratio analysis. At least 25 fields cells were analyzed per condition. One-way ANOVA statistical analysis and SEM are shown. (L) Representative images of WT and MTCH2 KO MEFs treated with non-targeting (NT) siRNA or with MFN2 siRNA, in control conditions, treated with HBSS for 4 h or treated with 10 μM CHX for 4 h. Mitochondria were labeled with MTDR. Scale bar 10 µm. (M) Classical classification of mitochondrial network morphology. Results are presented as average and SEM of three different experiments, and at least 30 cells were analyzed per condition. (N) Mitochondrial aspect ratio analysis. At least 25 fields were analyzed per condition. One-way ANOVA statistical analysis and SEM are shown. Data information: (C,F,K,N) ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001. Source data are available online for this figure.