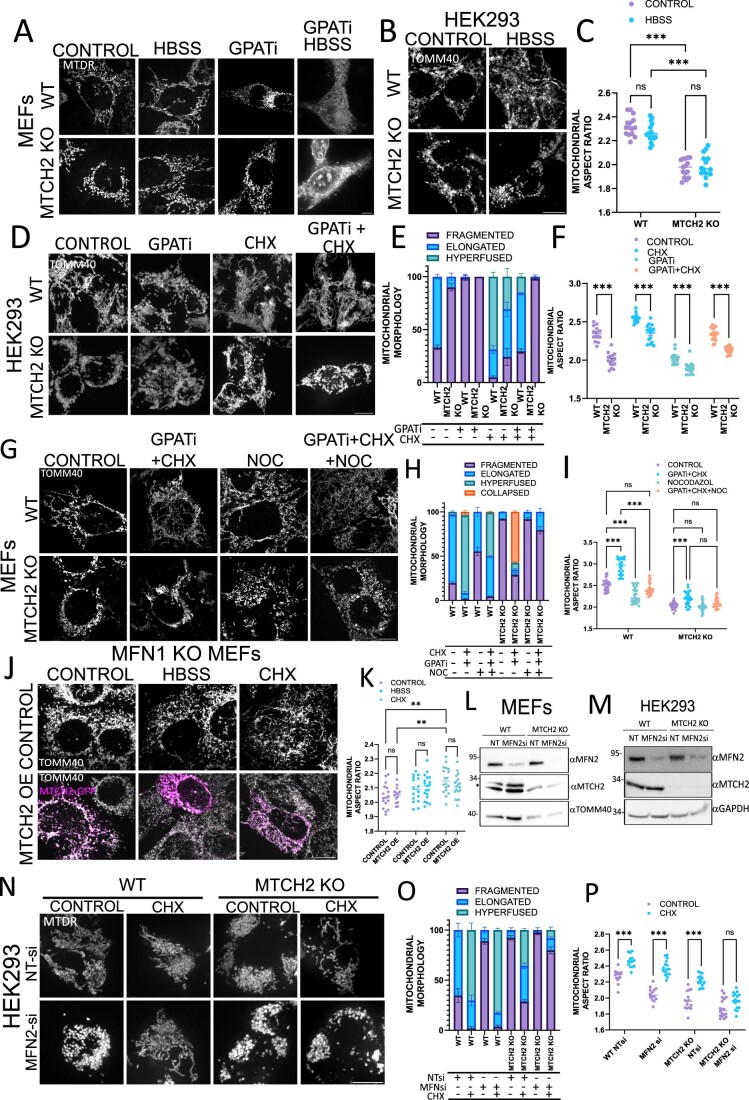
Figure EV4. MTCH2 cooperates with MFN2 and LPA synthesis to sustain mitochondrial fusion under cellular stress conditions.
(A) Representatives images of WT and MTCH2 KO MEFs either non-treated (control), treated with HBSS for 4 h, treated with GPATi for 16 h, or pretreated for 16 h with GPATi and then treated with HBSS for 4 h in the presence of the inhibitor. Mitochondria were labeled with MTDR. Scale bar 10 µm. (B) Representatives images of WT and MTCH2 KO HEK293T cells non-treated (control) or treated with HBSS for 4 h. Mitochondria were stained with αTOMM40 antibody. Scale bar 10 µm. (C) Mitochondrial aspect ratio analysis. At least 20 fields were analyzed per condition. One-way ANOVA statistical analysis. (D) Representatives images of WT and MTCH2 KO HEK293T cells either non-treated (control), treated with GPATi for 16 h, treated with 10 µM CHX for 4 h or pretreated for 16 h with GPATi and then treated with 10 µM CHX for 4 h in the presence of the inhibitor. Scale bar 10 µm. (E) Classical classification of mitochondrial network morphology. Results are presented as average and SEM of three different experiments, and at least 30 cells analyzed per condition. (F) Mitochondrial aspect ratio analysis. At least 20 fields were analyzed per condition. One-way ANOVA statistical analysis and SEM are shown. (G) Representatives images of WT and MTCH2 KO MEFs either non-treated (control), pretreated for 16 h with GPATi and then treated with 10 µM CHX for 4 h in the presence of the inhibitor, treated with 5 µM nocodazole (NOC) for 4 h or pretreated for 16 h with GPATi and then treated with 5 µM NOC and 10 µM CHX for 4 h in the presence of the inhibitor. Mitochondria were labeled with αTOMM40 antibody. Scale bar 10 µm. (H) Classical classification of mitochondrial network morphology. Results are presented as average and SEM of three different experiments, and at least 30 cells analyzed per condition. (I) Mitochondrial aspect ratio analysis. At least 25 fields were analyzed per condition. One-way ANOVA statistical analysis and SEM are shown. (J) Representatives images of MFN1 KO MEFs control or overexpressing MTCH2-GFP in control conditions or treated for 4 h with HBSS or CHX. Mitochondria were stained with αTOMM40 antibody. Scale bar 10 µm. (K) Mitochondrial aspect ratio analysis. At least 20 cells were analyzed per condition. One-way ANOVA statistical analysis and SEM are shown. (L) Western blot analysis of MFN2 in WT and MTCH2 KO MEFs treated with NT-siRNA or with siRNA targeting MFN2 expression, and TOMM40 serves as loading control. *Depicts a non-specific band. (M) Western blot analysis of MFN2 in WT and MTCH2 KO HEK293T cells treated with non-targeting (NT) siRNA or with MFN2 siRNA. GAPDH serves as a loading control. (N) Representative images of WT and MTCH2 KO HEK293T cells treated with non-targeting (NT) siRNA or with MFN2 siRNA non-treated or treated with CHX for 4 h. Mitochondria were labeled with MTDR. Scale bar 10 µm. (O) Classical classification of mitochondrial network morphology. Results are presented as average and SEM of three different experiments, and at least 30 cells analyzed per condition. (P) Mitochondrial aspect ratio. At least 20 fields were analyzed per condition. One-way ANOVA statistical analysis and SEM are shown. Data information: (C,F,I,K,P) ns, not significant, **P < 0.01, ***P < 0.001. Source data are available online for this figure.