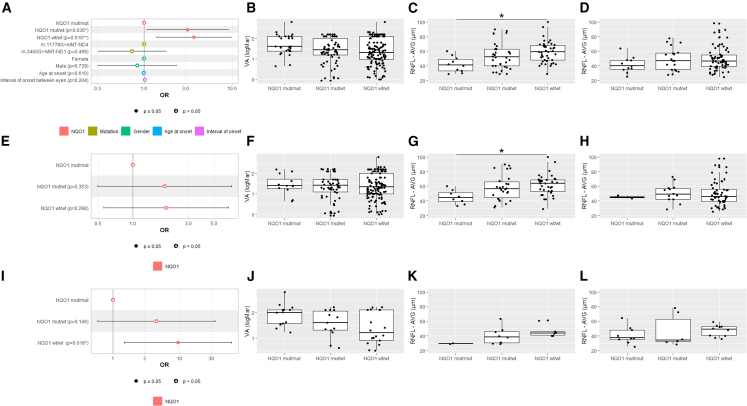
Figure 6.
NQO1 variants leading to very low protein levels match the IDB responder/non-responder analysis
(A, E, and I) Forest plots of odds ratios (ORs) for the binary visual outcome (responder/non-responder to IDB therapy).
(A) ORs calculated by multivariable GEE modeling on the 118 LHON patients treated with IDB therapy.
(E) ORs calculated by univariable GEE modeling on the LHON patients treated with IDB therapy carrying the m.11778G>A/MT-ND4 mutation.
(I) ORs calculated by univariable GEE modeling on the LHON patients treated with IDB therapy carrying the m.3460G>A/MT-ND1 mutation.
(B, F, and J) Boxplots of VA at last visit, with solid lines representing median values for NQO1 mut/mut, NQO1 mut/WT, and NQO1 WT/WT genotypes of all 118 LHON patients (B), LHON patients carrying the m.11778G>A/MT-ND4 (F), and LHON patients carrying m.3460G>A/MT-ND1 (J).
(C, G, and K) All Italian LHON patients (C), Italian LHON patients with m.11778G>A/MT-ND4 (G), and Italian LHON patients with m.3460G>A/MT-ND1 (K). Shown are boxplots of average RNFL thickness at last visit, with solid lines representing median values for NQO1 mut/mut, NQO1 mut/WT, and NQO1 WT/WT genotypes. All of these patients were evaluated with swept-source OCT (DRI Triton OCT, Topcon, Tokyo, Japan). ∗p < 0.05.
(D, H, and L) All German LHON patients (D), German LHON patients with m.11778G>A/MT-ND4 (H), and German LHON patients with m.3460G>A/MT-ND1 (L). Shown are boxplots of average RNFL thickness at last visit, with solid lines representing median values for NQO1 mut/mut, NQO1 mut/WT, and NQO1 WT/WT genotypes. All of these patients were evaluated with spectral-domain OCT (Heidelberg Spectralis OCT, Heidelberg Engineering, Heidelberg, Germany).