Figure 3.
GemCis ± atezolizumab versus GemCarbo ± atezolizumab induces proinflammatory transcriptional programs across multiple immune cell subsets in PBMCs
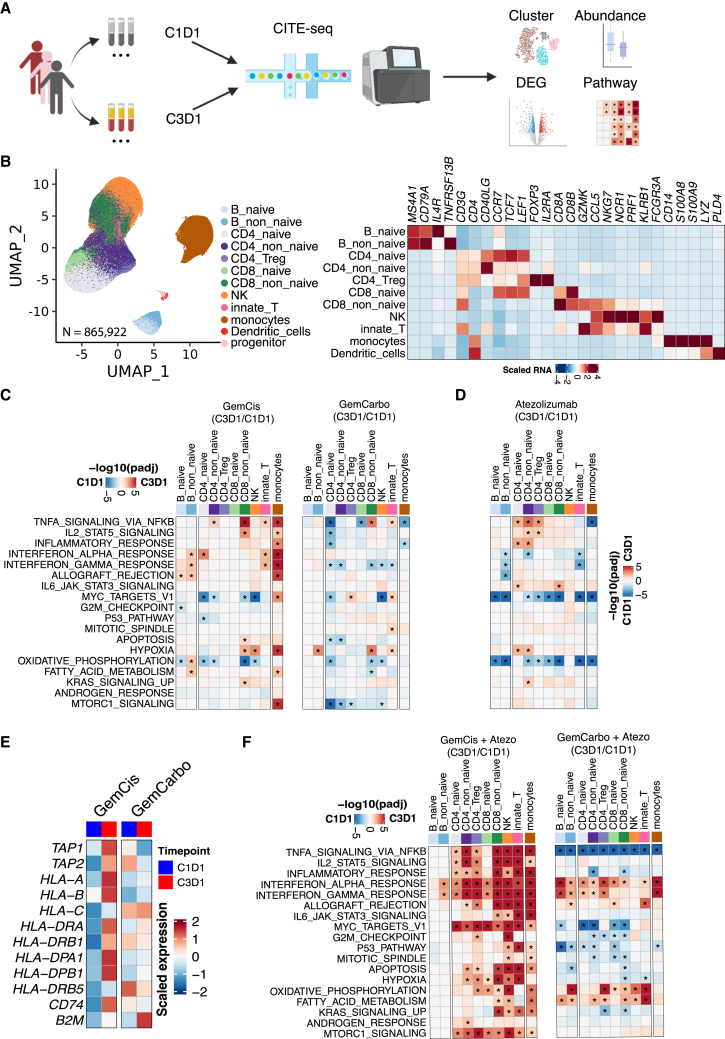
(A) Single-cell CITE-seq experimental design and analysis workflow.
(B) Uniform Manifold Approximation and Projection (UMAP) visualization of single cells captured, colored by major cell types (n = 865,922) (left), and heatmap showing scaled expression of the canonical marker genes across cell types (right).
(C and D) Heatmaps showing pathway enrichment on-treatment (C3D1) versus at baseline (C1D1) across multiple immune cell types. Treatments received were GemCis (left, n = 17 pairs) or GemCarbo (right, n = 16 pairs) in arm C (C) and atezolizumab (n = 33 pairs) in arm B (D).
(E) Comparison of MHC class gene expression on-treatment (C3D1) versus at baseline (C1D1) in monocytes collected from patients receiving GemCis or GemCarbo in arm C.
(F) Heatmaps showing pathway enrichment on-treatment (C3D1) versus at baseline (C1D1) across multiple immune cell types. Treatments received were GemCis + atezolizumab (left, n = 14 pairs) or GemCarbo + atezolizumab (right, n = 24 pairs) in arm A. In (C), (D), and (F), red indicates enrichment in on-treatment samples and blue indicates enrichment at baseline. The hue represents the false discovery rate (FDR) significance derived from the fgsea package. Black asterisks represent FDR < 0.05. See also Figures S3 and S4.