Figure 1.
Determination of malignant cell groups
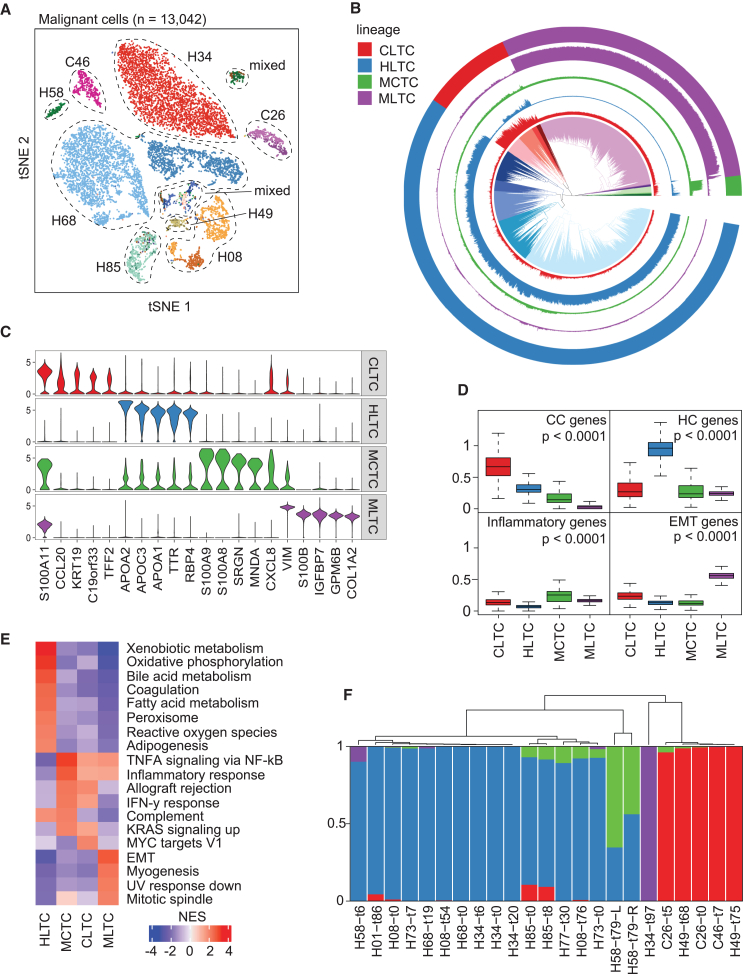
(A) t-SNE plot of all 13,042 malignant cells. Cells are assigned a general color by patient and a shade of that color by sample, with the lightest shade at baseline and successively darker hues for post-treatment samples. Patient IDs start with H and C to denote the clinical diagnosis of HCC and iCCA, respectively.
(B) Hierarchical relationship of all malignant cells. Inner section shows the hierarchical relationship among all cells, with colors showing the top 20 clades. Outer ring denotes the top four clades of the hierarchical tree as tumor lineages. The four interior rings indicate with matched colors the expression of the differentially expressed genes for each of the four major lineages.
(C) Violin plots of the expression of the top five differentially expressed genes by log fold change for each tumor lineage.
(D) Average expression of representative genes specific to cholangiocytes (CC), hepatocytes (HC), inflammation, or epithelial-mesenchymal transition (EMT) in malignant cells of each tumor lineage. The p values were calculated using a two-sided t test.
(E) Enriched pathways of each tumor lineage. Normalized enrichment score (NES) is shown for each pathway.
(F) Tumor lineage composition in each tumor sample with at least 15 malignant cells detected. Bars are colored by lineage: CLTC in red, HLTC in blue, MCTC in green, and MLTC in purple. See also Figures S1 and S2 and Tables S1 and S2.