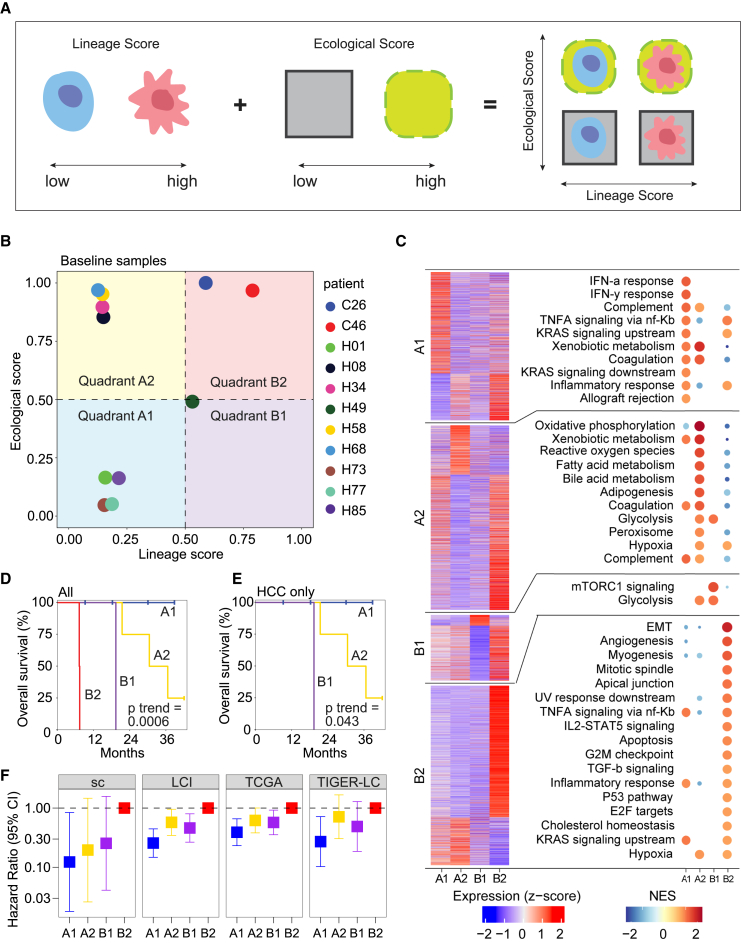
Figure 3.
Modeling a tumor ecosystem using lineage and ecological scores
(A) Graphical representation of using lineage and ecological scores to model a tumor ecosystem.
(B) Projection of the baseline samples from each of the 11 patients in the discovery single-cell cohort to the lineage-ecological space.
(C) Biological features of the tumors in each quadrant of the lineage-ecological space. The heatmap on the left shows the average expression of the top positive and negative differentially expressed genes in a quadrant. Each section of the heatmap, denoting differentially expressed genes for a quadrant (from top to bottom A1, A2, B1, and B2), has a corresponding plot on the right showing the top significantly enriched hallmark pathways for those genes. Pathways are ordered by high to low normalized enrichment score (NES). Only upregulated pathways are shown here.
(D and E) Kaplan-Meier plots of the 11 liver cancer patients (D) or only HCC patients (E) from the quadrants in (B). The p value was calculated using the log rank test for trend.
(F) Forest plot of hazard ratio in an additional HCC/iCCA single-cell cohort (sc) (n = 20) and three bulk HCC cohorts of LCI (n = 239), TCGA (n = 363), and TIGER-LC (n = 62). Quadrant B2 was used as the reference group for hazard ratio calculation. Bars show 95% confidence interval. See also Figures S5 and S6 and Tables S4 and S5.