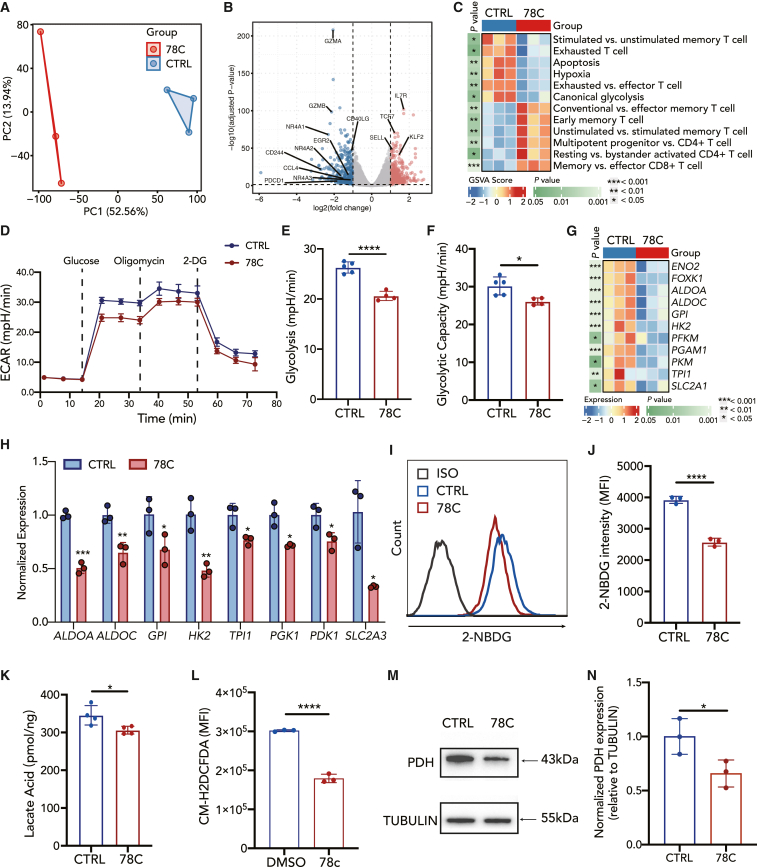
Figure 4.
CD38 inhibition results in reduced glycolysis metabolism in CAR-T cells
(A) Principal component analysis of CAR-T cells in different groups.
(B) Volcano plot illustrating differential gene expression analysis in CD38-inhibited CAR-T compared to control CAR-T cells after coculture with NALM6 cells at 1:1 (E:T).
(C) Heatmap of selected pathways enriched in genes significantly upregulated or downregulated in 78C-treated CAR-T cells. A single sample enrichment score was calculated for each pathway, and the mean was taken per response group. A color gradient ranging from blue to red indicates the mean normalized enrichment score (ranging from −2 to +2) of pathways enriched in induced (red) or repressed (blue) genes.
(D–F) Metabolic rate as measured by Seahorse analysis of extracellular acidification rate (ECAR) of control (n = 5 technical replicates) or CD38-inhibited (n = 4 technical replicates) CAR-T cells after coculture with NALM6 cells.
(G) Heatmap of differentially expressed genes in canonical glycolysis pathway in comparison with the control group.
(H) mRNA level of glycolysis-related transcription factors in control or CD38-inhibited CAR-T cells after coculture with NALM6 cells. Data are mean ± SD of 3 technical replicates.
(I) Flow cytometric analysis of 2-NBDG uptake in each group.
(J) Median fluorescence intensity of 2-NBDG in control or 78C-treated CAR-T cells after coculture with NALM6 cells. Data are mean ± SD of 3 independent experiments from three different donors.
(K) Cellular lactate acid level in control or 78C-treated CAR-T cells after coculture with NALM6 cells (n = 4 technical replicates from two different donors).
(L) Flow cytometric analysis cytoplasmic ROS in control or 78C-treated CAR-T cells after coculture with NALM6 cells by CM-H2DCFDA staining (n = 3 biological replicates).
(M and N) Western blot analysis of PDH (M) and normalized PDH expression relative to TUBULIN in each group (N). NALM6-stimulated CAR-T cells were treated with DMSO/78C for 3 days. Quantitative analysis of western blot data obtained in n = 3 technical replicates is shown, normalized to tubulin.