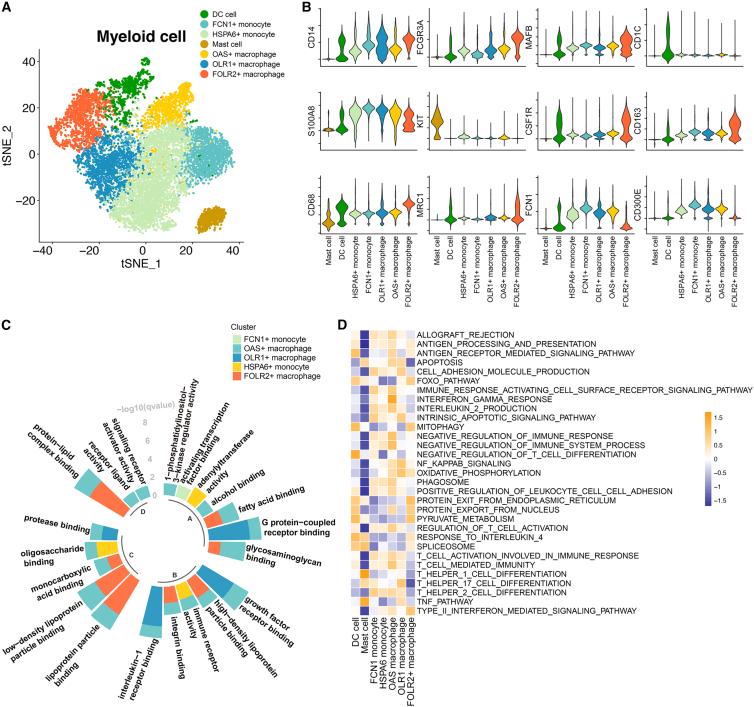
Figure 2.
The properties of myeloid subpopulations
(A) The t-distributed stochastic neighbor embedding (t-SNE) shows seven subclusters of myeloid cells (patients, n = 26).
(B) Stacked violin plot showing the scaled expression level of markers for each population.
(C) The enriched Gene Ontology (GO) terms for each monocyte/macrophage population. The GO analysis was performed with the gene signature for each monocyte and macrophage subpopulation. The height of each column represents the –log10(p) value of enriched biological processes for each subpopulation.
(D) Heatmap showing the immune pathway activities in myeloid populations. Purple indicates low values, while yellow indicates high values.