Figure 3.
The anti-inflammatory properties of FOLR2+ macrophages
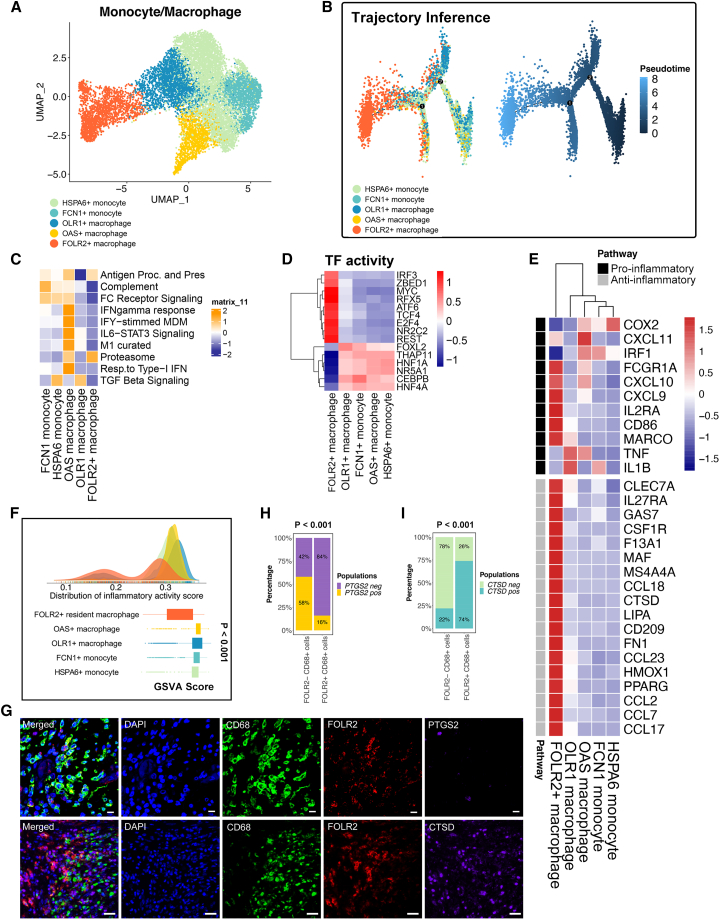
(A) The UMAP plot shows five monocyte/macrophage subpopulations (patients, n = 26).
(B) Pseudotime analysis of five subclusters of monocytes/macrophages (patients, n = 26). The color key from deep blue to light blue indicates the pseudotime score from low to high.
(C) Heatmap of the single-sample gene set enrichment analysis (ssGSEA) scores for gene sets in monocyte/macrophage subtypes, calculated according to single-cell RNA sequencing (scRNA-seq) data.
(D) Transcription factor (TF) activity in monocyte/macrophage subpopulations.
(E) Heatmap of the expression of M1-related and M2-related genes from the scRNA-seq data.
(F) Density plot indicating the density distribution of the inflammatory response activity score in monocyte and macrophage populations and boxplot indicating the inflammatory response activity score in monocyte and macrophage populations. The inflammatory response activity score was calculated with the ssGSEA method using the inflammatory response signature on the single-cell transcriptome. The p value was calculated with a Kruskal-Wallis test.
(G) Multiplex immunohistochemistry (mIHC) staining for CD68 (green), FOLR2 (red), PTGS2 (purple), CTSD (purple), and DAPI (blue). Scale bar, 10 μm. Biological replicates, n = 5.
(H) The percentage of FOLR2+ CD68+ cells and FOLR2− CD68+ cells expressing PTGS2. A total of 100 cells were included for analysis. Significance was evaluated by a chi-square test.
(I) The percentage of FOLR2+ CD68+ cells and FOLR2− CD68+ cells expressing CTSD. Significance was evaluated by a chi-squared test.