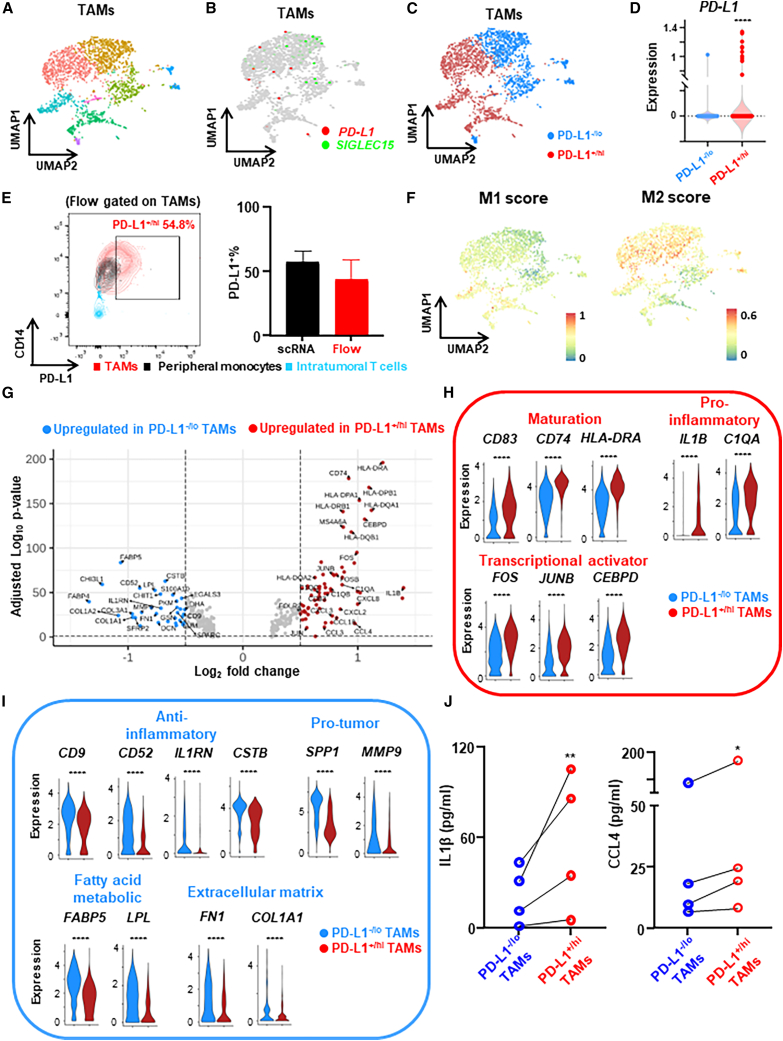
Figure 1.
Expression profile differences between PD-L1+/hi vs. PD-L1–/lo TAMs from human breast tumors revealed by scRNA-seq
(A) scRNA-seq analysis of TAMs (n = 2,220 cells) from untreated primary breast tumor (n = 5 patients, ER+) shown as a UMAP, highlighting identified clusters.
(B) UMAP showing mutually exclusive expression of PD-L1 and SIGLEC15 in TAMs.
(C) Dichotomization of TAM clusters into PD-L1+/hi and PD-L1–/lo subpopulations.
(D) PD-L1 expression of PD-L1+/hi and PD-L1–/lo subpopulations.
(E) Representative flow cytometry plot showing PD-L1+% TAMs, peripheral monocytes, and intratumoral T cells from the same tumor tissue used for scRNA-seq (left panel), and PD-L1+% quantification was compared between analysis via scRNA-seq and flow cytometry (right panel).
(F) Overlay of the expression of common M1 and M2 signature genes in the PD-L1+/hi vs. PD-L1–/lo TAM dichotomization.
(G) Volcano plot showing differentially expressed genes (DEGs) between PD-L1+/hi vs. PD-L1–/lo TAMs.
(H and I) Expression distribution of selected genes involved in maturation, pro-inflammatory or transcriptional activator (H), and anti-inflammatory, pro-tumor, fatty acid metabolic, or extracellular matrix (I) between PD-L1+/hi and PD-L1–/lo TAMs. ∗∗∗∗p < 0.0001. Wilcoxon rank-sum test.
(J) PD-L1+/hi and PD-L1–/lo TAMs were flow sorted from freshly prepared single-cell suspension of digested breast tumor (n = 4) and supernatants were collected for ELISA after 16 h. Paired t test. ∗p < 0.05, ∗∗p < 0.01.