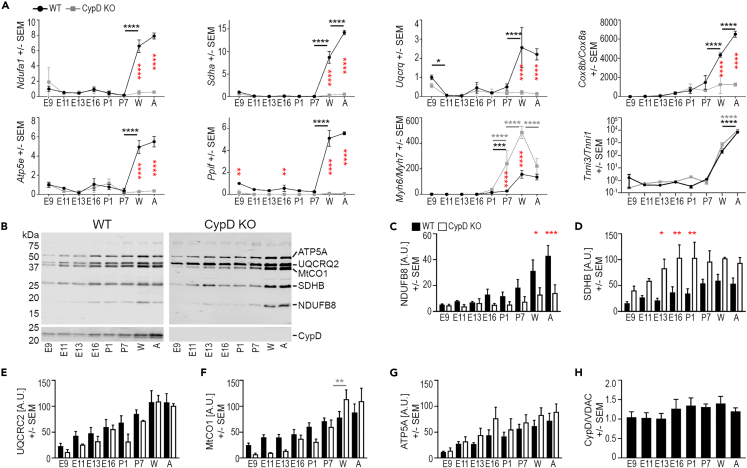
Figure 2.
CypD deletion alters mitochondrial gene and protein expression in the developing heart
(A) qRT-PCR analyses of mitochondrial (Ndufa1, Sdha, Uqcrq, Cox8a, Cox8b, Atp5e, and Ppif) and contractile apparatus (Myh6, Myh7, Tnni1, Tnni3) gene expression. Data for Cox8b/a, Myh6/7, and Tnni3/1 are expressed as ratios and all data was normalized to the expression of Polr2A.
(B–G) Representative immunoblot images (B) and graphs of the expression of NDUFB6 (C), SDHB (D), UQCRC2 (E), MtCO1 (F), and ATP5A (G) and CypD (H) demonstrate changes in proteins levels during development in tissue homogenates. Data from immunoblots for ETC proteins (C-G) was normalized to protein loading (A.U., arbitrary units) and not VDAC labeling due to data in Figure 1L. Note: an additional lane of adult homogenate in the top WT panel was removed from between E13.5 and E16.5 for clarity.
(H) CypD protein expression normalized to VDAC expression is stable during cardiac development in WT hearts. Data is presented as Mean ± SEM and analyzed by one-way ANOVA with Tukey’s or Sidak’s multiple comparison test to compare data between WT and CypD KO cells at each age (red stars in A) and also between successive ages of the same genotype (WT-underlined black stars, CypD KO-underlined gray stars). Only significant differences noted, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; N: A = 3–6, C-H = 3 hearts/age and genotype.