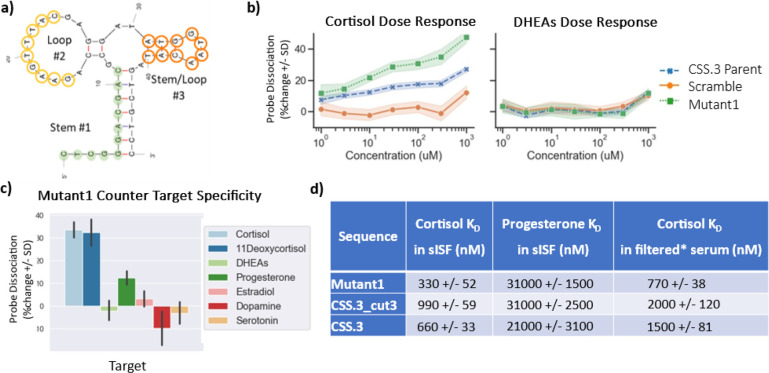
Figure 4.
Sequence optimization of the CSS.3 aptamer for an improved binding affinity. (a) Regions selected for nucleotide randomization in loop #2 (yellow) and stem and loop #3 (orange) are highlighted. (b) Dose–response curves showing increased probe dissociation for mutant compared to CSS.3 parent sequence and scrambled negative control sequence in the presence of cortisol (left) but not DHEAs (right). (c) Specificity of optimized mutant showing strong response to cortisol and 11-deoxycortisol, moderate response to progesterone and no response to other structurally similar targets. (d) DNA sequences were immobilized on BLI sensor tips through biotin and binding to cortisol was measured directly (without the use of complementary sequences), it is demonstrated that the selected mutant has 3-fold better binding affinity and selectivity against progesterone compared to the stem-optimized CSS.3_cut3 sequence. The mutant sequence also performs better in filtered human serum as a surrogate for testing in real ISF. (*) Serum filtered using 30 kDa MWCO filter.