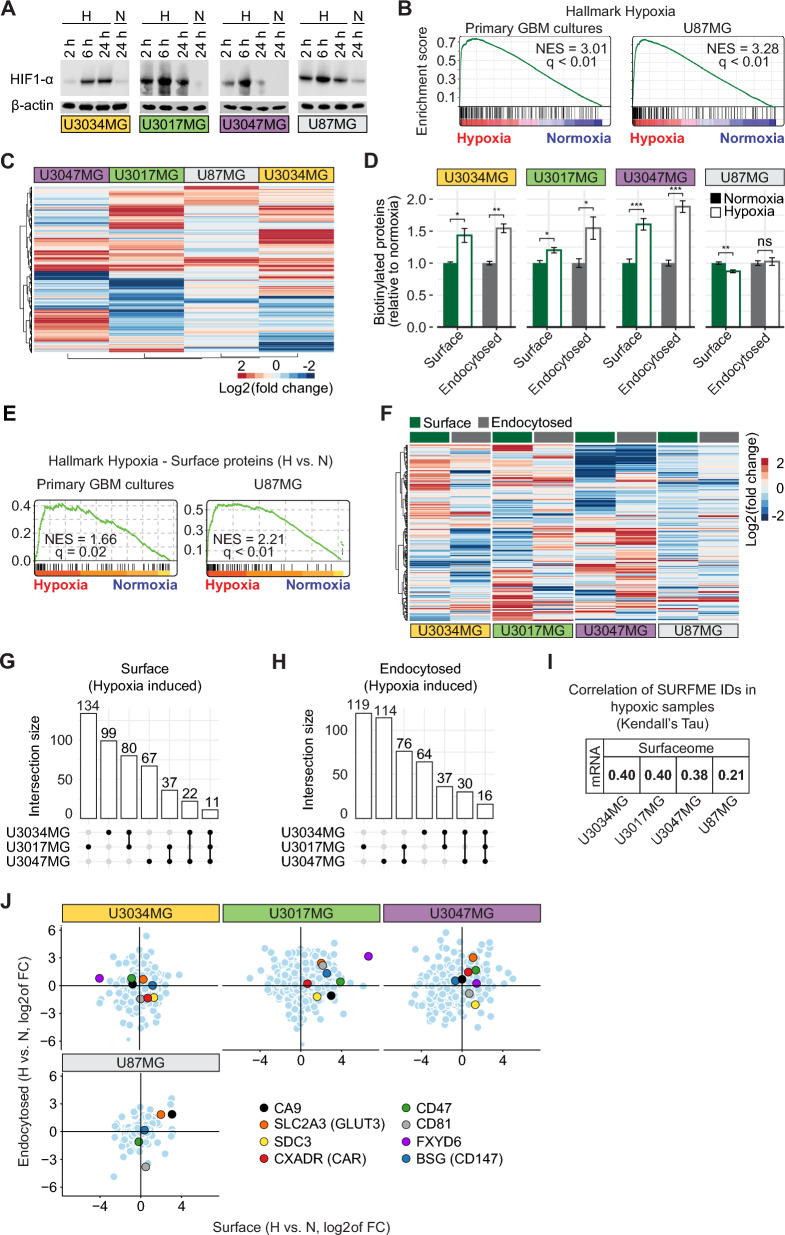
Fig. 4.
Hypoxic remodeling of the global surfaceome and endocytome in GBM cells. A Normoxic and hypoxic cell lysates at the indicated time-points were probed for HIF1-α by immunoblotting, with β-actin as loading control. B GSEA shows a significant overrepresentation of hallmark hypoxia-induced genes in mRNA of hypoxic vs. normoxic primary GBM (left) and U87MG (right) cells. C Heatmap of SURFME filtered mRNA data shows highly divergent hypoxic regulation between GBM cell types. D FACS quantification of global biotinylated surfaceome and endocytome in normoxic and hypoxic GBM cell cultures, as indicated. Data were normalized for normoxia (set at 1) and are expressed as the mean fold ± SD from three independent experiments, each performed in triplicates. *, **, *** P < 0.05, 0.01, and 0.001, respectively; ns, not significant. E GSEA on the surfaceome of hypoxic primary GBM (left) and U87MG (right) cells displays a significant enrichment of known hypoxia-responsive proteins when compared to the respective normoxic cells. F Hierarchical clustering of surface and endocytosed proteins identified in hypoxic compared to normoxic primary GBM and U87MG cells. G, H Upset plots show the distribution of common and unique hypoxia-induced surfaceome (G) and endocytome (H) proteins in GBM cells. I Kendall’s Tau correlation of hypoxia-induced IDs indicates weak associations between SURFME-filtered mRNA and protein abundance. J SURFME proteins identified in the surfaceome as well as the endocytome of GBM cells, ranked according to log2 of the fold change of hypoxia vs. normoxia and divided into quadrants. Top-right, proteins with higher surface abundance and more efficient endocytic capacity in hypoxic conditions