Fig. 4.
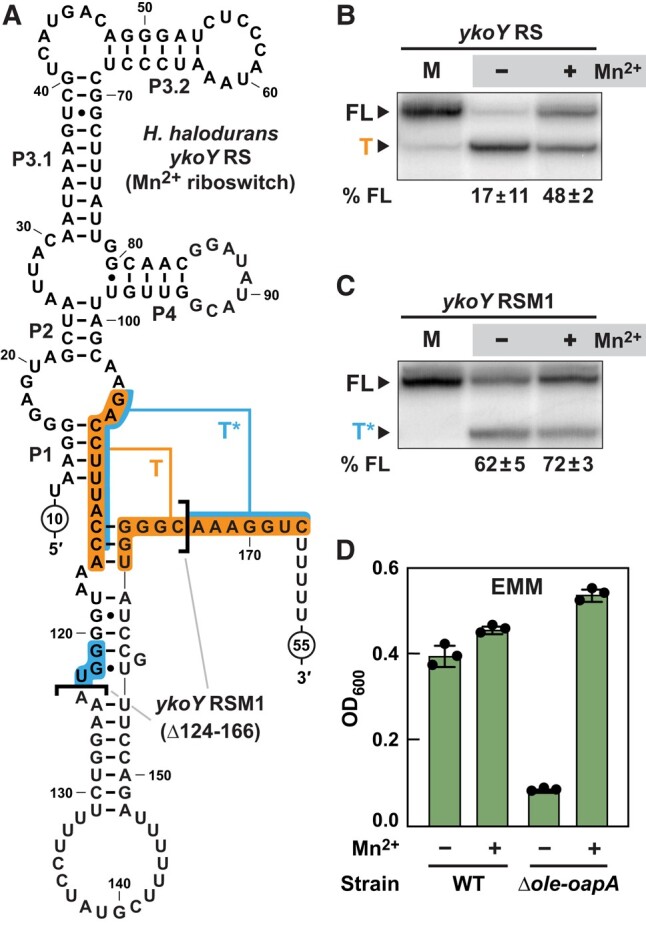
Δole-oapA suppressor isolates carrying a 43-base-pair deletion upstream of the ykoY gene disrupt a Mn2+ riboswitch to yield increased YkoY production. A) Sequence and secondary structure model for the H. halodurans ykoY riboswitch in its predicted Mn2+-bound state (genetic “ON” conformation). Brackets identify the 43-nucleotide RNA region deleted in the RSM1 mutation, which is a deletion of nucleotides 124 through 166. Indicated are the natural intrinsic terminator stem (T) and the predicted fortuitous terminator (T*) formed by the RSM1 suppressor mutant. B) Transcription termination assays using the WT ykoY riboswitch sequence (ykoY RS). FL, full-length transcript; T, terminated transcript at the T terminator; (‒), no supplemented Mn2+; (+), 200 µM supplemented MnCl2. % FL represents the average and standard deviation of FL transcripts based on three replicate experiments. C) Transcription termination assay using the ykoY RSM1 riboswitch identified in Δole-oapA suppressor selection in EMM. Annotations are as described for (B), except that T* designates the product band resulting from termination at the T* terminator. D) Growth assays in EMM with H. halodurans WT and Δole-oapA strains in the absence (−) or presence (+) of 10 µM MnCl2. Cells were grown for 48 h. Bars represent the average from three biological replicates wherein each replicate was comprised of three technical replicates. Error bars represent standard deviation.
