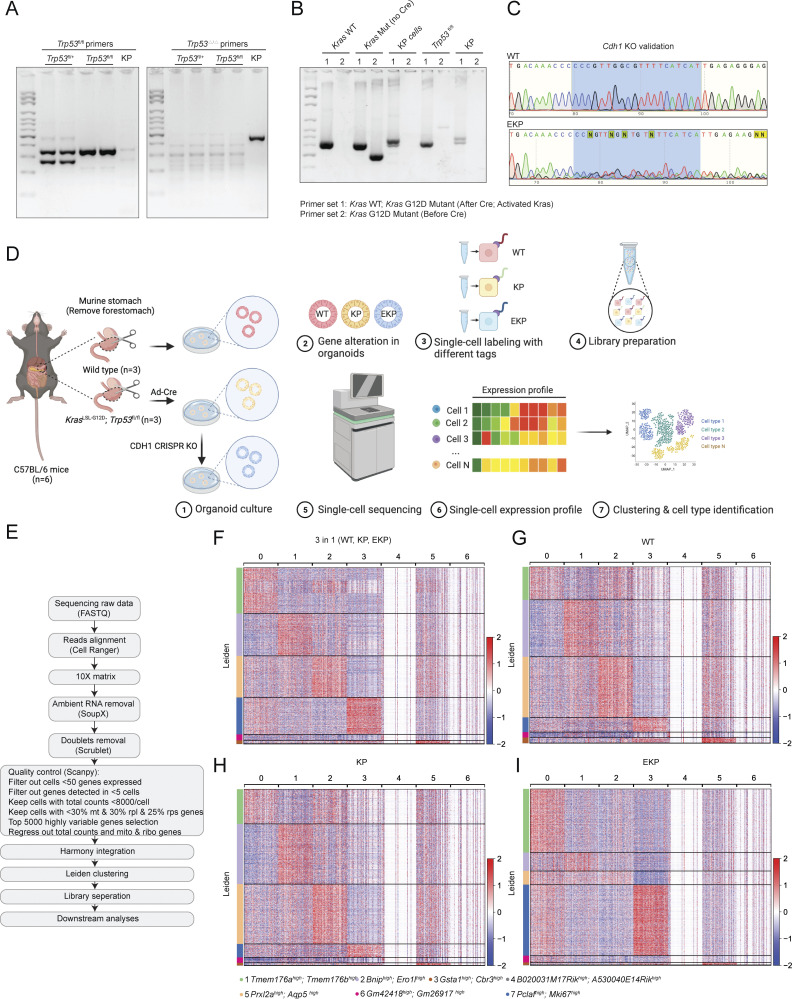
Figure S4.
Validation of genetic engineering and scRNA-seq analysis of mouse GOs. (A–C) Genotyping results of KP organoids (A and B). After adeno-Cre treatment, KP organoids lost Trp53, while KrasG12D was activated in KP organoids. After CDH1 CRISPR KO, we performed Sanger sequencing to compare the sequence of CDH1 in WT and EKP (C). The five targeting sequences against CDH1 are shown in CRISPR/Cas9-based gene knockout in GOs. The primers used for genotyping are shown in Table S2. (D) Illustration of the workflow for stomach tissue collection and dissociation, gene manipulation of the gastric organoids (GOs), sample preparation of multiplex scRNA sequencing. (E) Workflow of single-cell library preparation. (F) Heatmap of each cell clusters of integrated datasets, including WT, KP, and EKP. (G–I) Separate heatmap of each cell clusters of WT, KP, and EKP datasets, respectively. All data are derived from two or more independent experiments with the indicated number of mice. Source data are available for this figure: SourceData FS4.