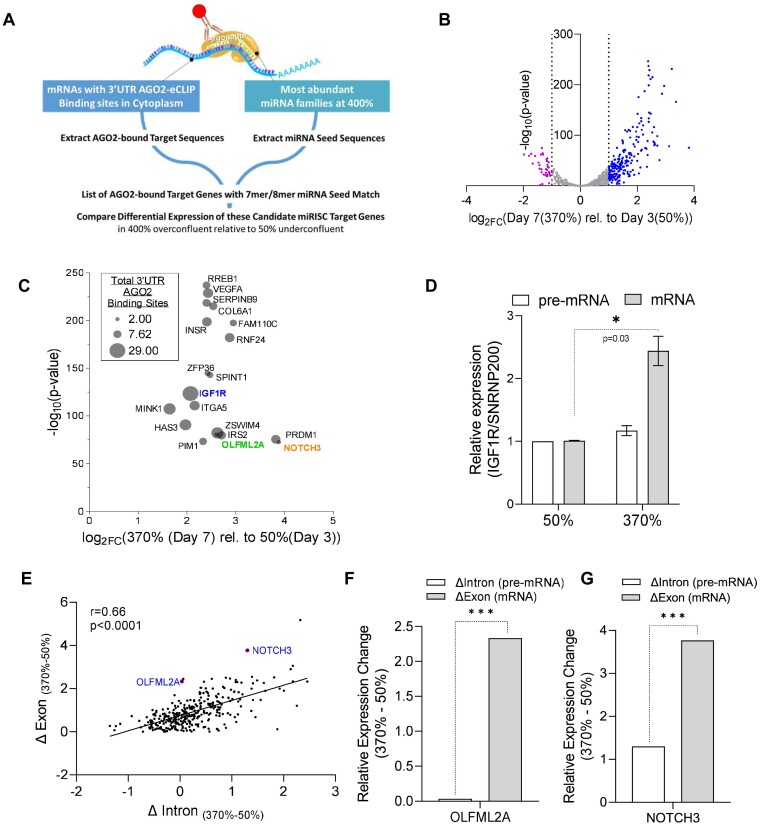
Figure 5.
Nuclear localization of AGO2 relieves repression of cytoplasmic 3′UTR targets. (A) Candidate refinement using AGO2-eCLIP sequencing, small-RNA-sequencing, miRNA family seed alignment, and whole transcriptome RNA-sequencing. (B) Volcano plot of differential expression from RNA-seq of candidates with abundant miRNA family seed matches to 3′UTR AGO2-eCLIP binding sites in the cytoplasm. (C) Plot showing differential expression, significance, and number of cytoplasmic AGO2-eCLIP binding sites within 3′UTR of candidate genes targeted by miRISC. (D) Basal expression level of pre-mRNA and mRNA for IGF1R measured with RT-qPCR from RNA harvested on Day 3 (underconfluent, 50%) and Day 7 (overconfluent, 370%). Significance denoted with *, P-value equals 0.03. (E) Exon-Intron Split Analysis (EISA) of AGO2-eCLIP candidates with sites of 3′UTR occupancy and abundant miRNA family seed matches. (F) EISA analysis showing changes in expression of intronic reads (pre-mRNA) and exonic reads (mRNA) at high density (370%) relative to low density (50%) for (F) OLFML2A, P-value = 3.32 × 10−11, FDR = 1.47 × 10−8 and (G) NOTCH3, P-value = 2.75 × 10−11, FDR = 1.37 × 10−8.