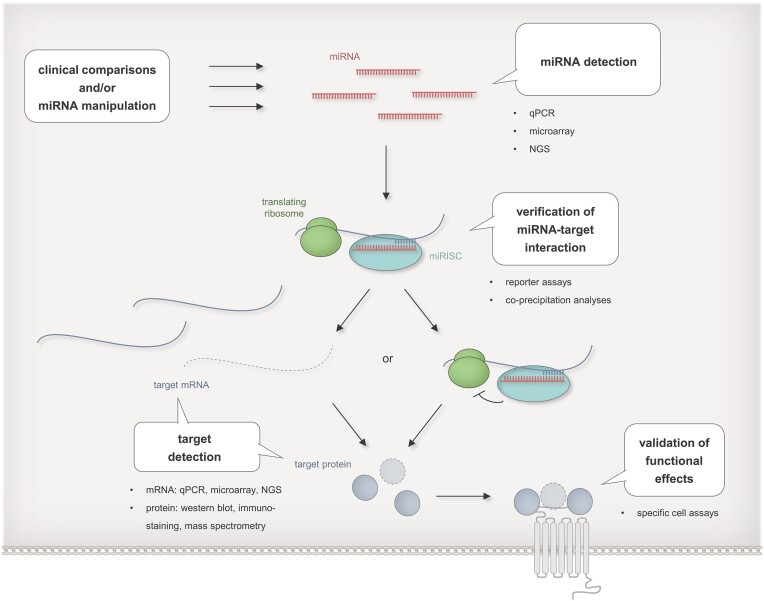
Figure 4.
Experimental strategies for the identification of miRNA targets. MiRNA targets can be identified by comparisons of e.g. healthy vs. diseased cells or by downstream analysis upon experimental miRNA manipulations. Differential abundances of miRNAs and target mRNA are detectable by qPCR, microarray analyses, or next generation sequencing (NGS), differential levels of target proteins by western blot, immunostaining or mass spectrometry, direct interaction between miRNAs and their target mRNA by reporter assays or co-precipitation analyses, and down-stream miRNA effects by proliferation assays, viability assays or by assays tailored for specific miRNA regulated cell functions. [MiRNAs are indicated in red, target mRNAs in blue, and target proteins as blue spherical bodies. Interactions between miRNA and mRNA are depicted by opposite comb-shaped lines. Degradation of mRNA is indicated by dashed lines and reduction in protein expression by dashed outlines. Functional effects are symbolized by a grey shaded receptor icon. Ribosomes are shown as green bodies, RISCs as turquoise bodies.]