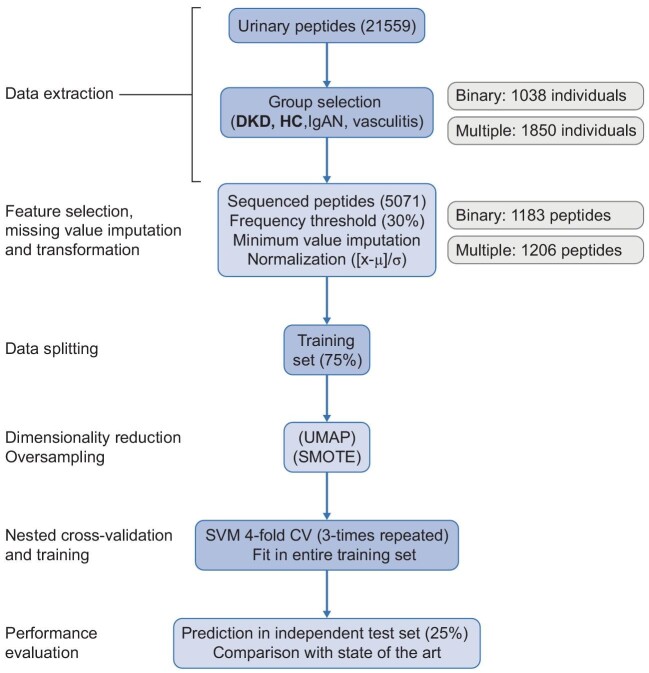
Figure 1:
Study design. The urinary peptide datasets of a cohort of 1850 HC and CKD (DKD, IgAN and vasculitis) individuals were implemented into a supervised machine learning pipeline for classification based on disease (or lack thereof). The pipeline was performed separately for DKD and HC classes (binary classification) as well as all classes (multiclass classification). Initially, a splitting of the classification data into a training (75%) and a test (25%) set was performed. Then, the sequenced peptides present in at least 30% of the respective participants, were considered for further analysis and normalized in the training and test sets {[x-mean(x)]/standard deviation(x), considering the training set} after missing peptide values of each dataset were imputed based on the respective minimum values. A dimensionality reduction with the UMAP algorithm was performed (or skipped), while as an additional step during the training procedures in the multiclass classification only, the oversampling algorithm SMOTE [31] was applied. The latter produced synthetic participants in all classes until a certain ratio of the (initially) majority class (i.e., IgAN) was achieved, so as to account for the class imbalance. During a three-times repeated four-fold CV, SVM models were trained (in three out of four folds of the training set) and their performance was recorded (on the remaining fold) along the lines of an iterative search that relied on a Bayesian optimization [35] of the hyperparameters. The model that achieved the highest average accuracy across all the CV folds was selected as having the optimal combination of hyperparameter values. Subsequently, the selected model was trained in the entire training set and then tested for its predictive accuracy in the independent test set. μ, feature mean; σ, feature standard deviation; SMOTE, Synthetic Minority Over-sampling Technique; CV, cross-validation in training set.