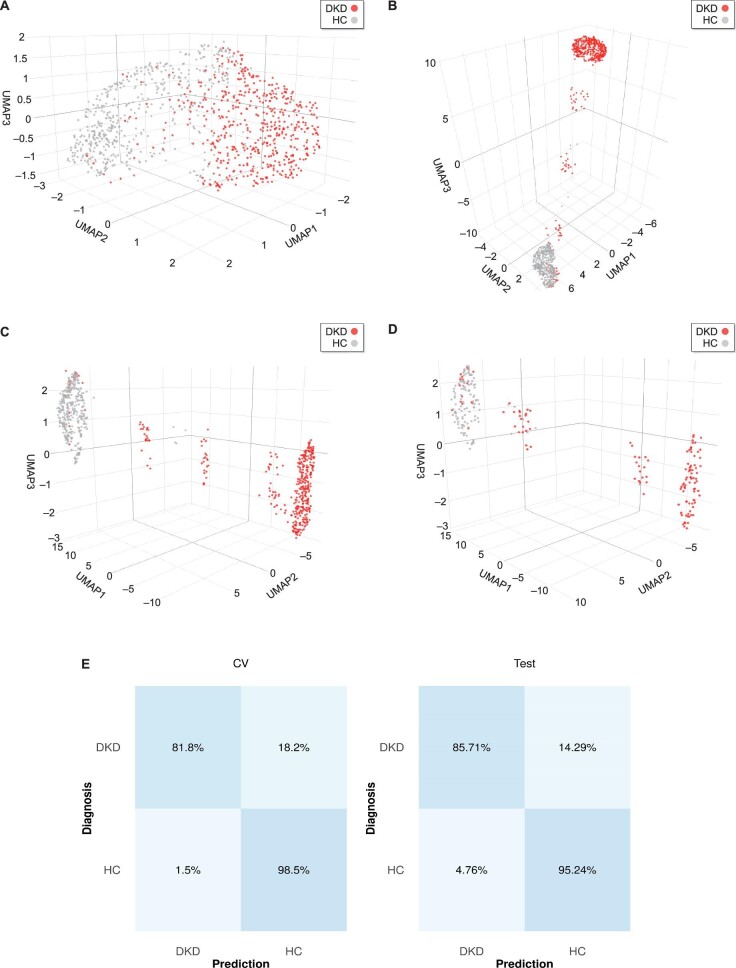
Figure 2:
Binary classification results. The peptidomic profiles of DKD (red) and HC (gray) participants were used as a basis for the default hyperparameters of the UMAP algorithm in its (A) unsupervised as well as (B) supervised version. Cluster formation was more evident when the supervised UMAP with tuned parameters was performed, as observed in the (C) training set and (D) independent test set embeddings. (E) Confusion matrices based on the results of the training set cross-validation (CV, average across all folds) as well as the predictions in the independent test set. Classification accuracies are displayed in percentages.