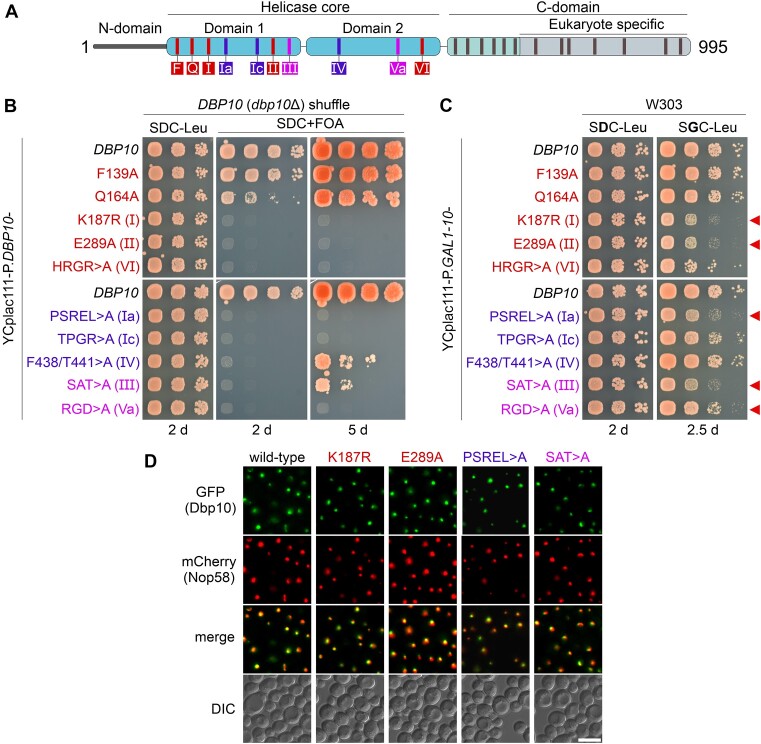
Figure 1.
Genetic analyses of catalytic dbp10 mutants. (A) Domain organization of the DEAD-box helicase Dbp10. Conserved motifs involved in ATP-binding/hydrolysis, RNA-binding, and communication between RNA-binding and ATP-binding motifs that were mutated in this study are indicated in red, blue, and pink, respectively. (B) A DBP10 (dbp10Δ) shuffle strain was transformed with plasmids harbouring wild-type DBP10 or indicated dbp10 mutant alleles under transcriptional control of the DBP10 promoter. Transformants were spotted in 10-fold serial dilutions on SDC-Leu or 5-FOA containing plates (SDC + FOA) and growth was monitored after incubation at 30 °C for the indicated days. (C) Over-expression of DBP10 and indicated dbp10 catalytic mutants under the control of the galactose-inducible GAL1-10 promoter. Transformants were spotted in 10-fold serial dilution on plates containing glucose (SDC-Leu; repressed condition) or galactose (SGC-Leu; induced condition) and growth was monitored after incubation at 30°C for the indicated days. Red arrows indicate dominant-negative dbp10 mutants. (D) The subcellular distribution of GFP-tagged wild-type Dbp10 and indicated catalytic Dpp10 mutant variants in yeast cells was monitored by fluorescence microscopy. Co-localization with the nucleolar marker Nop58-mCherrry revealed a predominant nucleolar protein localization of all tested Dbp10 variants. DIC: differential interference contrast. Scale bar is 5 μm.