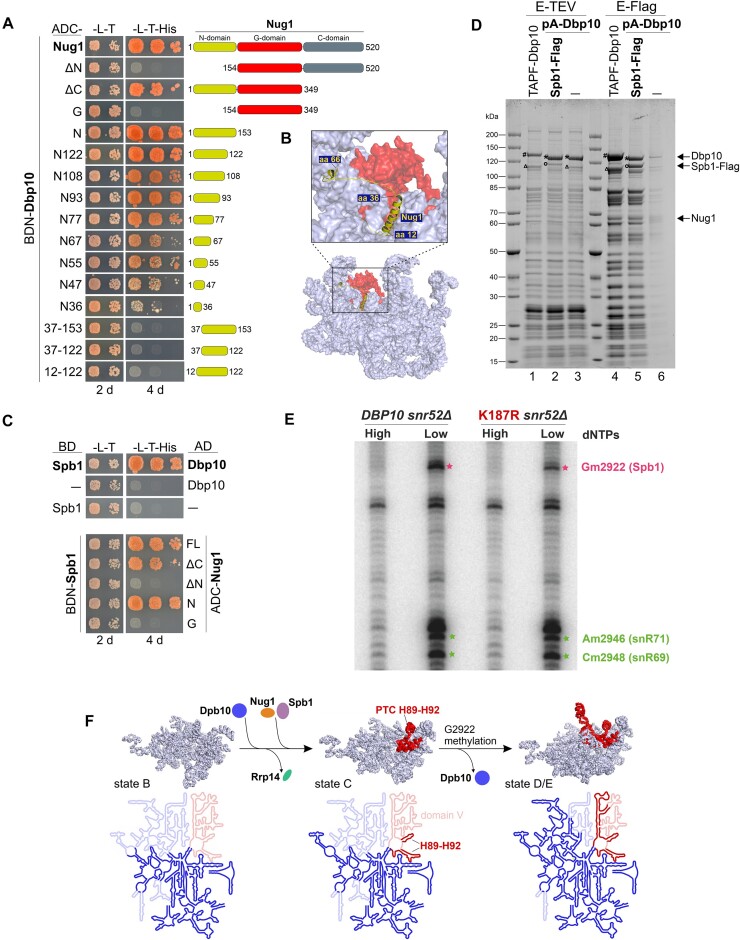
Figure 4.
Pre-ribosomal Dbp10–Nug1–Spb1 interaction and G2922 methylation by the Spb1 methyltransferase. (A) Indicated Nug1 constructs fused to the Gal4 activation domain (AD) were tested for a yeast two-hybrid interaction with Dbp10 fused to the Gal4 DNA-binding domain (BD). Cells were spotted in 10-fold serial dilutions on SDC-Leu-Trp (-L-T) and SDC-Leu-Trp-His (-L-T-His) plates and growth was monitored upon incubation at 30°C for two and four days, respectively (B) The interacting N-terminal protein part of Nug1 (yellow) resolved on a nucleoplasmic pre-60S cryo-EM structure (PDB 6YLH). The 25S rRNA is shown as surface representation with PTC rRNA helices H89-H92 coloured in red. (C) Spb1 fused to the Gal4 DNA-binding domain (BD) was tested for a Y2H interaction with Dbp10 (upper panel) and indicated Nug1 constructs (lower panel) fused to the Gal4 activation domain (AD). Cells were spotted in 10-fold serial dilutions on SDC-Leu-Trp (-L-T) and SDC-Leu-Trp-His (-L-T-His) plates and growth was monitored upon incubation at 30°C for the indicated days. (D) Split-tag affinity purification via endogenous proteinA-TEV-Dbp10 (1st purification step) and plasmid-borne Spb1-Flag (2nd purification step). The TEV (proteinA) eluate (lane 2) and Flag eluate (lane 5) were analysed by SDS page and Coomassie staining. As comparison a TAP-Flag-Dbp10 construct (lanes 1 and 4) and proteinA-TEV-Dbp10 alone (lanes 3 and 6; negative control) were used for affinity purifications. Relevant proteins are indicated: #Cbp-Flag-Dbp10, *Dbp10 (after removal of tag by TEV protease), °Spb1-Flag, ΔSpb1 (untagged). (E) Primer extension analyses using RNAs co-purified with Nsa1-FTpA from cells lacking the snR52 snoRNA and expressing DBP10 WT or the dbp10 K187R mutant. Primer extensions were performed using AMV reverse transcriptase at high (1 mM) or low (0.01 mM) dNTP concentration to detect and quantify the methylation levels of G2922 and two neighbouring nucleotides, A2946 and C2948, the ribose of which are methylated by the box C/D snoRNPs snR71 and snR69, respectively. (F) Dbp10-dependent Nug1/Spb1 assembly and PTC H89-H92 rRNA folding. Upper panel: the pre-rRNA (including 25S, 5.8S, and ITS2) of state B (PDB 6EM4), state C (PDB 6EM1), and state E (PDB 6ELZ) pre-60S cryo-EM structures is shown in light blue with 25S domain V rRNA coloured in red. The proposed assembly stage of Dbp10/Nug1/Spb1 to these nucleolar pre-60S intermediates and Rrp14 release, as well as the stage of G2922 methylation are indicated. Lower panel: the 25S rRNA folding state within the depicted particles is illustrated schematically. Already folded/visible rRNA regions are coloured in bright blue/red, unfolded/invisible rRNA regions in faint blue/red. The 25S domain V rRNA including the H89-H92 Dbp10 target is coloured in red.