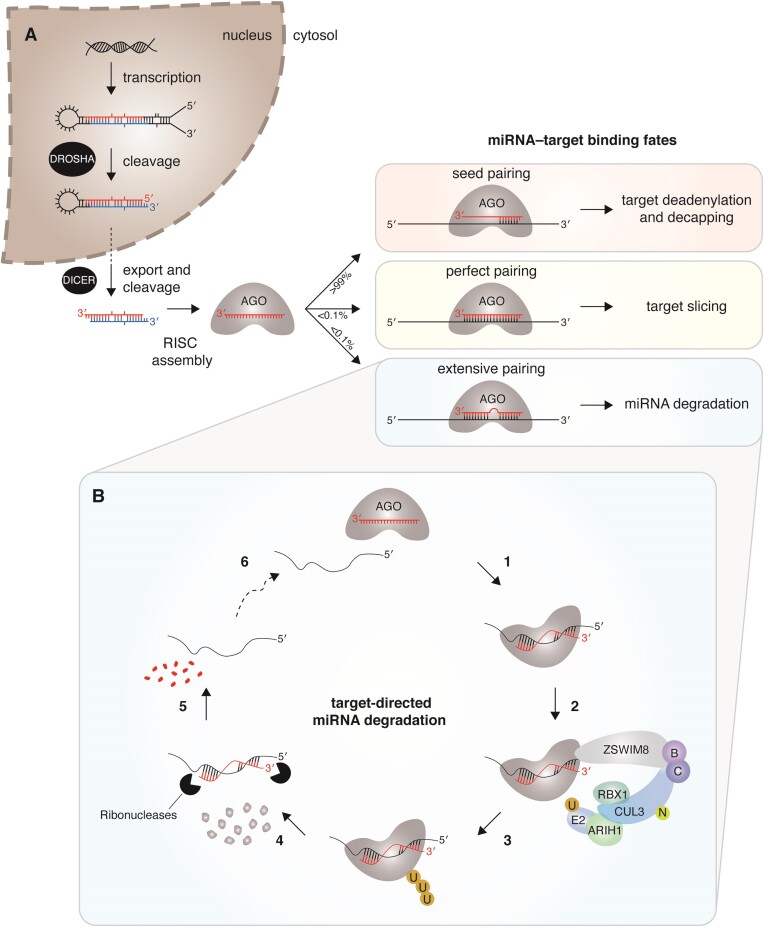
Figure 1.
The miRNA life cycle. (A) miRNA biogenesis and targeting are depicted. After sequential processing in the nucleus and cytosol by DROSHA/Microprocessor and DICER, respectively, the miRNA duplex is loaded into the AGO effector protein and one strand is removed. The fate of the AGO–miRNA complex and target RNA depend on the extent of base-pairing between miRNA and target. These outcomes include target degradation via deadenylation and decapping, target degradation via endonucleolytic cleavage (also known as slicing), and miRNA degradation. (B) In the current model of target-directed miRNA degradation, extensive complementarity between the AGO–miRNA complex and target RNA induces a stabilized ternary complex (1), which recruits the ZSWIM8 substrate adapter, other subunits of the E3 ubiquitin ligase including neddylated (N) CUL3, ELONGINS B and C, ARIH1, and RBX1, and an ubiquitin-conjugating enzyme (E2) (2), leading to polyubiquitination (U) (3) and proteasomal degradation of AGO (4), followed by degradation of the deprotected miRNA (5). No longer bound to miRNA, the target RNA can interact with a new AGO–miRNA complex (6), initiating another round of miRNA degradation.