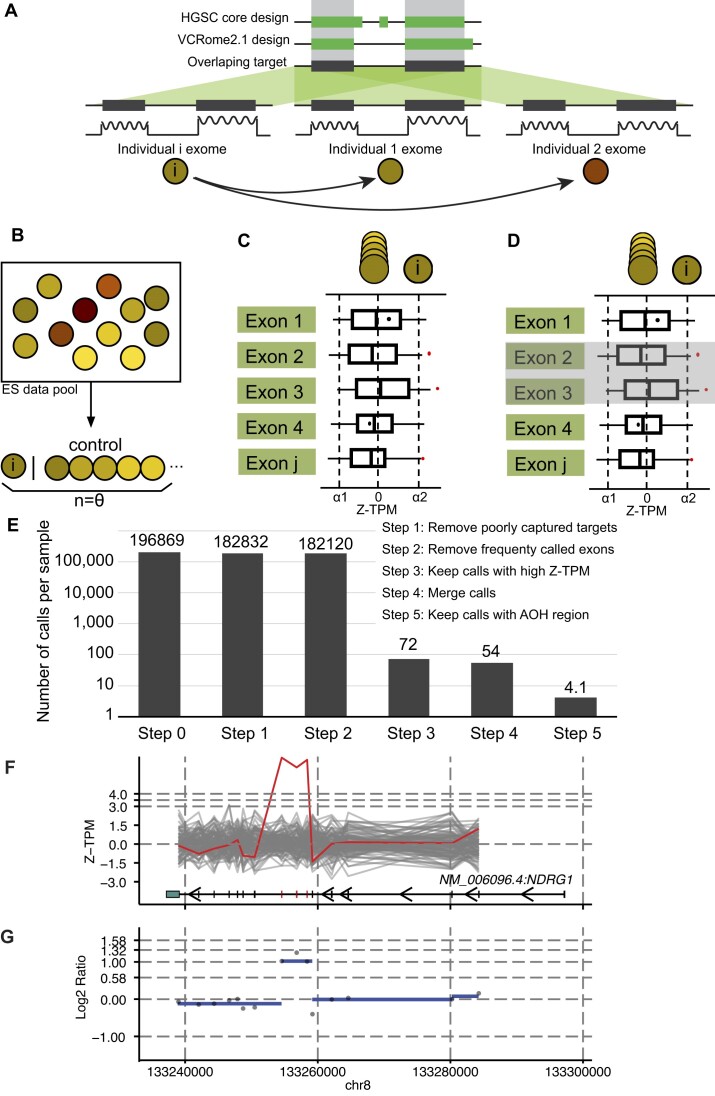
Figure 1.
The computational framework for HMZDupFinder. (A–D) The HMZDupFinder workflow demonstrates the harmonization of exome capture design, nearest reference selection, Z-TPM normalization, and homozygous duplication enrichment process. (E) Estimated numbers of candidate CNVs after each ES data computational step. (F, G) An example of combined visualization of Z-TPM and log2 ratio validating a homozygous intragenic duplication. (F) The line plot displays Z-TPM values for exons that match the annotations from the MANE transcript of the investigated gene, which is shown at the bottom. A red, jagged horizontal line signifies the value from the subject, with grey lines representing the closest reference samples. The segmented log2 ratio of the TPM value in the subject is illustrated as a blue dashed line in panel (G).