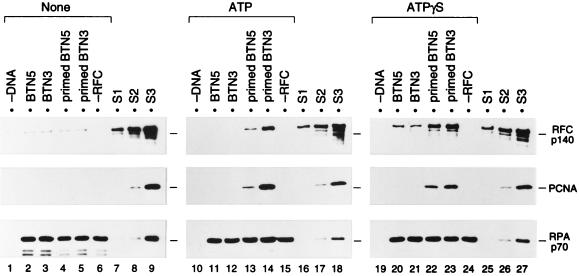
FIG. 3.
ATP- and primer-specific formation of a primer recognition complex on DNA fixed onto agarose beads. The loading reaction was carried out in the absence of nucleotide (lanes 1 to 6) or in the presence (lanes 10 to 15) of ATP or ATPγS (lanes 19 to 24), using 1 pmol each of BTN5 without primer (lanes 2, 11, and 20), BTN3 without primer (lanes 3, 12, and 21), primed BTN5 (lanes 4, 13, and 22), or primed BTN3 (lanes 5, 14, and 23), along with 2 pmol of RPA, 2.2 pmol of RFC, and 20 pmol of PCNA trimer. Reactions without DNA (lanes 1, 10, and 19) or RFC (lanes 6, 15, and 24) were also performed. The beads, after incubation for 30 min at room temperature, were washed at 0°C with buffer A containing no nucleotide (lanes 1 to 6), 0.2 mM ATP (lanes 10 to 15), or 0.2 mM ATPγS (lanes 19 to 24). The bound proteins and standards were analyzed as described in the legend to Fig. 2. Each set of samples (lanes 1 to 9, 10 to 18, or 19 to 27) was run on the same SDS-polyacrylamide gel. Note that although 10-fold more PCNA was used here as well as for the study shown in Fig. 4, the efficiency of loading of PCNA was essentially the same with either amount of PCNA. S1, S2, and S3, standards (see legend to Fig. 2).