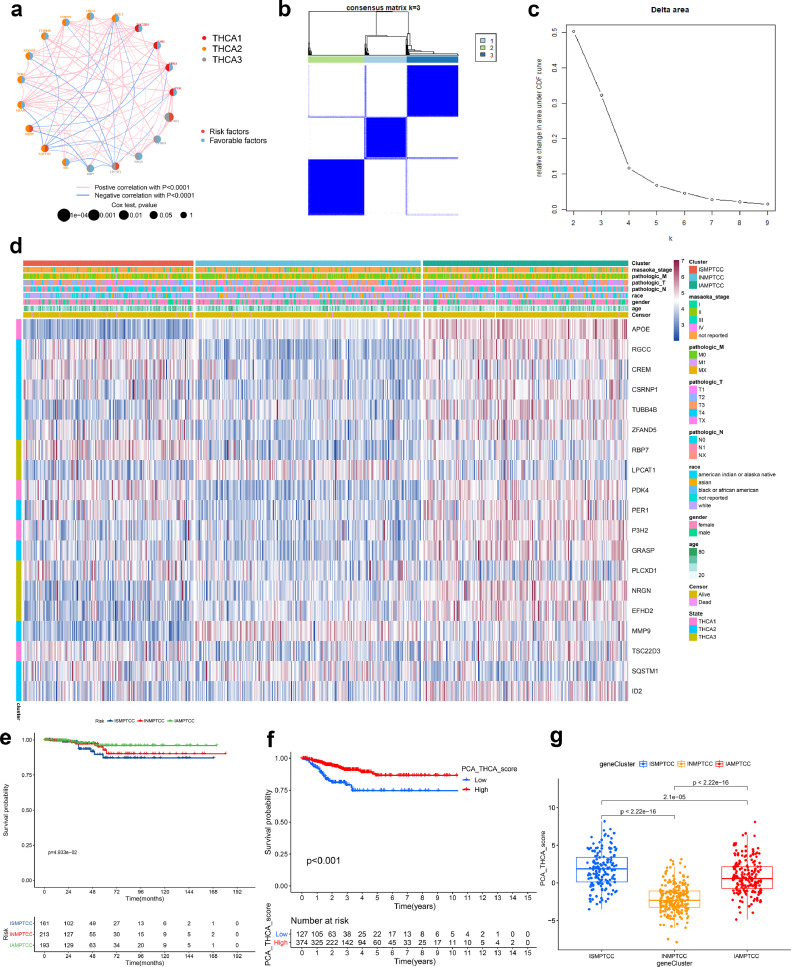
Fig. 4.
Identification of MD&PRGs and IMPTCC construction
(a) Co-expression network analysis elucidating the co-expression relationships of 19 MD&PRGs across three cellular differentiation states.
(b-c) Bivariate clustering results from consistency matrices suggest optimal clustering into three groups at k = 3, evident from pronounced inter-group differences, minimal intra-group heterogeneity, and an expansive area under the CDF curve. Considering clustering distinction and maximal curve area principles, k = 3 is deemed optimal.
(d) The heatmap displays the differential expression of 19 MD&PRGs across three IMPTCC clusters and three cellular differentiation states, correlating with their respective clinical characteristics.
(e) Kaplan-Meier survival curves contrasting survival outcomes of the three clusters.
(f) Kaplan-Meier survival curves comparing survival outcomes between high and low PCA groups.
(g) Box plot delineating PCA score variations among the three clusters.