Abstract
Fog causes enhancement of bacterial loading in the atmosphere. Current study represents the impact of occurrences of fog on the alteration of diversity of airborne bacteria and their network computed from metagenomic data of airborne samples collected at Arthauli (25.95°N, 85.10°E) situated at central Indo-Gangetic Plain (IGP) during 1–14 January 2021. A distinct bacterial diversity with a complex network is identified in foggy condition due to the enrichment of unique types of bacteria. Present investigation highlights a statistically significant enrichment of airborne pathogenic bacteria found in a unique ecosystem within air evolved due to the occurrences of fog over central IGP. In the foggy network, Cutibacterium, an opportunistic pathogen, is identified to be interacting maximum (21 edges) with other bacteria with statistically significant copresence relation, which are responsible for various infections for human beings. A 40–60% increase (p < 0.01) in the abundance of pathogenic bacteria for respiratory and skin diseases is noticed in fog period. Among the fog-enriched bacteria, Cutibacterium, Herbaspirillum, Paenibacillus, and Tsukamurella are examples of opportunistic bacteria causing various respiratory diseases, while Paenibacillus can even cause skin cancer and acute lymphoblastic leukemia.
Keywords: Fog, Airborne bacteria, Pathogens, Diversity, Indo-Gangetic plain
1. Introduction
In the recent era, the study of bioaerosols (bacteria, fungi, viruses, and other living or dead microorganisms) demands more attention because they contribute about 25% to total atmospheric aerosols and up to 80% to the atmospheric particles’ sizes larger than 1 μm [1]. It also has an adverse impact on all kinds of ecosystems, climate, and human society [2]. The role of meteorological conditions in the survival of these airborne microorganisms is entirely mysterious. However, extreme weather conditions like fog, cyclones, and dust make the scenario more complex.
Bioaerosols cannot survive alone for a longer duration in hot, sunny atmospheric conditions because dry air having high temperature with low relative humidity (RH) and high solar radition is not favorable for their sustenance. A few studies reported that high abundances of bioaerosols emitted during dust storms can travel long distances [3,4]. An unmanned aircraft measurement reported that smoke plumes from wildland fires over the western United States contained about 4-fold increased cell counts with respect to the background air [5]. In addition, foggy days enhance airborne microbial community due to favorable atmospheric conditions providing plenty of water droplets in low temperatures and low solar radiation [[6], [7], [8]]. Saikh and Das, 2023 reported that bioaerosol loading in a relatively drier and warmer atmosphere drastically decreases on a typical normal day than in foggy conditions. Fog acts as a medium for microbial growth by providing nutrients and protecting them from desiccation, oligotrophic conditions, and harmful incoming solar radiation [9,10].
The bioaerosol communities are not only harmful to life but also strongly recognized as highly diverse, ranging from non-viable to dormant to actively metabolized. A few are identified with an enormous impact on human health. Airborne microbes like Bacillus anthracis and Burkholderia cepacia can potentially cause many diseases like respiratory tract infections and urinary tract infections (UTI) [2,11,12]. Exposure to very low infectious bioaerosols can cause devastating human diseases. In earlier studies, it was reported that a single organism of Francisella tularensis can act as a contagious dose to humans, and a few cells of Mycobacterium tuberculosis are enough to overcome average lung clearance and inactivation mechanisms in a susceptible host [13]. It is worth mentioning here that Cutibacterium, Mycobacterium, Paenibacillus, and Williamsia are some examples of pathogenic airborne bacteria that causes various health issues in human. Cutibacterium causes breast infection, infective endocarditis, skin abscesses, and device-related infections [14,15]. Mycobacterium causes tuberculosis, initiated by the deposition of Mycobacterium tuberculosis on the lungs' alveoli from aerosol droplets [16,17]. Paenibacillus is responsible for chronic kidney disease, sickle cell disease, premature birth, Whipple's disease, hydrocephalus, skin cancer, chronic interstitial nephropathy, and acute lymphoblastic leukemia [18]. Williamsia infection includes pulmonary infections, bacteremia, endophthalmitis, and perinatal sepsis [[19], [20], [21], [22]]. The study of bioaerosols is becoming more relevant in many disciplines, including public health, environmental microbiology, industrial and agricultural engineering, and biological warfare.
Fog is a very common atmospheric phenomenon in Northern India, occurring frequently in winter season over IGP [[23], [24], [25]]. Almost half of the winter season experiences fog over North-western part of IGP [25]. Occurrences of fog significantly impact microbial loading [7,8,26]. The present study aims to investigate the alteration of airborne bacterial diversity due to occurrences of fog over central IGP and their effect on human health. The novelty of the current research work is to report a distinct fog-induced airborne bacterial diversity with a complex network comparing with pre-fog and post-fog conditions and establish that a foggy atmosphere over central IGP is suitable for the survival of pathogenic bacteria, which have relatively higher abundances and are responsible for different human diseases.
2. Materials and methodology
2.1. Site description
Airborne samples were collected from 1st to January 14, 2021 round the clock at Arthauli (25.95°N, 85.10°E), a rural site in Vaishali district of North-Western Bihar located at central IGP. The sampling period was selected to coincide with the most probable season of fog occurrence (Dec–Jan) over IGP [[23], [24], [25]]. The samples were collected at a height of 10 m above the ground, on the roof of a residential house surrounded by croplands, and in a remote place away from large industries. This sampling location is unique for collecting fog samples because no metropolitan or heavy industrial area was nearby, which could significantly contribute to local anthropogenic sources. The selected sampling site is located on the east bank of Gandak River, and the mainstream of Ganga River flows about 50 km south of the site. In addition, plenty of brick chimneys are present along the coast of Ganga River, mainly releasing smoke in the evening. Also, in the evening, the local villagers set fire to dry leaves and branches to keep themselves warm in harsh winter conditions, which may contribute to the total abundance of microbes.
Fig. 1(a and c) are satellite images obtained from Meteorological and Oceanographic Satellite Data Archival Centre (MOSDAC), Space Applications Centre, Indian Space Research Organisation (ISRO) on 10th and January 12, 2021 at 0300 h, respectively. These images are captured by INSAT 3D satellite. The location of occurrences of fog (red color) is shown in Fig. 1(a and c). The sampling site is marked by a yellow box over the satellite-captured images. However, such fog intensity does not appear over the sampling site, as shown in Fig. 1(a), i.e., on January 10, 2021. Fig. 1(c) shows the occurrences of fog over sampling site. Fig. 1(b and d) are the photos taken from the rooftop of the sampling site on 10th and January 12, 2021, respectively. A clear view is noticed in Fig. 1(b), whereas in Fig. 1(d), dense fog is noticed with visibility significantly reduced.
Fig. 1.
INSAT-3D satellite images captured occurrences of fog over IGP. The box (yellow) presents location of sampling site over central IGP. (a) Satellite image on January 10, 2021 at 0300 h showing sampling site free from fog occurred over North-western IGP (b) Photo taken from the rooftop of sampling site on January 10, 2021 present typical clear winter day. (c) Occurrences of fog over sampling site on January 12, 2021 at 0300 h depicting high intensity of fog spreading from west to east IGP, (d) Photo taken on January 12, 2021 at same location, indicating low visibility in a typical foggy day.
2.2. Meteorological conditions
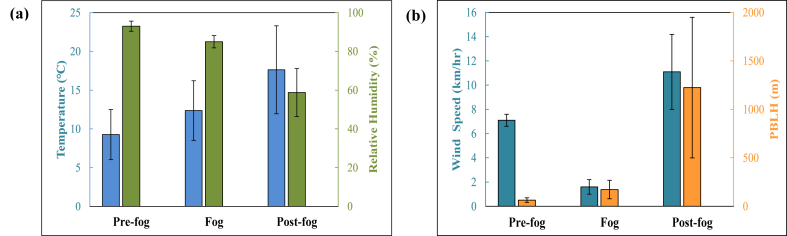
An Automated Weather Station (AWS) was installed at the rooftop of sampling site to collect hourly meteorological parameters like RH, temperature, and wind speed (WS). In addition, Planetary Boundary Layer Height (PBLH) data were retrieved from the National Centre for Medium Range Weather Forecasting (NCMRWF), Ministry of Earth Sciences (https://rds.ncmrwf.gov.in/). These meteorological parameters were used to classify the atmospheric conditions into three groups: pre-fog, fog, and post-fog. Fig. 2 shows the mean and standard deviation of temperature and RH [Fig. 2(a)], and WS and PBLH [Fig. 2(b)]. During pre-fog condition temperature, RH, WS, and PBLH were observed to be 9.3 ± 3.2 °C, 93.0 ± 2.6 %, 7.1 ± 0.5 km/h, and, 63.4 ± 23.1 m, respectively, 12.4 ± 3.8 °C, 85.0 ± 3.2 %, 1.6 ± 0.6 km/h, and, 172.1 ± 95.6 m, respectively under foggy condition, and, 17.6 ± 5.7 °C, 58.8 ± 12.4 %, 11.1 ± 3.1 km/h, and, 1224.7 ± 724.9 m, respectively under post-fog condition.
Fig. 2.
Variation of (a) temperature and RH, and (b) WS and PBLH in pre-fog, fog and post-fog.
2.3. Computation of back-trajectories
Computation of 3-days air parcel back-trajectories over sampling site was done using Hybrid Single-Particle Lagrangian Integrated Trajectory (HySPLIT) model (version-4) to identify the air travel path and a possible source region. Back-trajectories were computed for two days, i.e., 10 and January 12, 2021, at an initial arrival altitude of 100 m, as shown in Fig. 3. NCEP-NCAR (National Centers for Environmental Prediction-National Center for Atmospheric Research) reanalysis data were used to retrieve spatial distribution of wind vectors (at 950 hPa), RH at 0.995 sigma level, and air temperature at 2 m.
Fig. 3.
Spatial distribution of RH, temperature (level 2 m; open contours) with wind speed and direction over the Indian subcontinent region on a typical winter day (10th January) retrieved from NCEP-NCAR Reanalysis data. Three-days air parcel back-trajectories on 10th (clear day) and 12th January (foggy day) using HySplit model at an initial height of 100 m at sampling site marked by a star (blue) are shown. A slower movement of wind on foggy day originated from west IGP has been noticed by back-trajectory analysis.
2.4. Sample collection
Air samples were collected continuously for 4 h on sterile 4.7 cm filters of 0.22 μm pore size (MF-Millipore Mixed Cellulose Ester Membrane, Merck Life Science Private Limited, India), using an air suction pump. The pump was stopped for 1 h between two successive sample collections to prevent overheating. Filter papers were transferred into cryovials containing 3 ml of 50 mM:50 mM Tris: EDTA buffer (TE buffer, pH-7.8). Notably, nine fog events were observed during the campaign period; however, only five were considered for the present study due to overlapping atmospheric conditions during sample collection period. Details of the samples collected under different conditions are mentioned in Table 1.
Table 1.
Summary of samples collected under different atmospheric conditions.
| Atmospheric Condition | Sample name | Sampling Date (dd/mmm/yyyy) | Sampling Time | Raw read | Valid read | OTU Number | Genus Number |
|---|---|---|---|---|---|---|---|
| Pre-fog | V14 | Jan 11, 2021 | 02:30 a.m.–06:30 a.m. | 315,286 | 307,362 | 1012 | 134 |
| V19 | Jan 12, 2021 | 02:30 a.m.–06:30 a.m. | 443,510 | 438,188 | 2245 | 135 | |
| V29 | Jan 14, 2021 | 02:30 a.m.–06:30 a.m. | 191,065 | 183,602 | 867 | 109 | |
| Fog | V10 | Jan 10, 2021 | 07:00 a.m.–11:00 a.m. | 839,784 | 821,241 | 3659 | 189 |
| V15 | Jan 11, 2021 | 07:00 a.m.–11:00 a.m. | 666,945 | 652,609 | 4038 | 226 | |
| V20 | Jan 12, 2021 | 07:00 a.m.–11:00 a.m. | 362,627 | 356,079 | 3122 | 159 | |
| V23 | Jan 12-13, 2021 | 10:30 p.m.–02:30 am | 411,715 | 400,691 | 3419 | 226 | |
| V24 | Jan 13, 2021 | 02:30 a.m.–06:30 a.m. | 272,399 | 266,727 | 2741 | 202 | |
| Post-fog | V11 | Jan 10, 2021 | 12:00 p.m.–04:00 p.m. | 192,124 | 190,019 | 626 | 76 |
| V16 | Jan 11, 2021 | 12:00 p.m.–04:00 p.m. | 278,524 | 269,735 | 1259 | 149 | |
| V21 | Jan 12, 2021 | 12:00 p.m.–04:00 p.m. | 179,078 | 175,859 | 1050 | 119 | |
| V25 | Jan 13, 2021 | 07:00 a.m.–11:00 a.m. | 117,349 | 114,745 | 672 | 140 |
2.5. DNA isolation
Microbial cell suspension was prepared following the protocol mentioned in the literatures [8,27]. Invitrogen PureLink™ Genomic DNA kit (ThermoFisher Scientific, USA) was used to extract DNA from the samples, according to the manufacturer's protocol, after cell suspension preparation. Air samples yield low concentrations of DNA (<30 ng) even after the amplification of V3 regions of 16S rRNA, which is insufficient for sequencing [27]. Contamination from kit reagents, which may contain DNA traces of bacteria (Kitome), may impact the low-concentration microbiome analysis [28,29]. So, kitome was isolated from the kit without cell suspension, and the reads obtained were eliminated from the samples to acquire actual airborne bacterial diversity.
2.6. Amplification of 16S rRNA gene fragments, sequencing of V3 regions
V3 regions of 16S rRNA genes were PCR amplified with “Fusion Primer Protocol” using universal forward primer (341f- 5′-CC TAC GGG AGG CAG CAG- 3′) and universal reverse primer (515r- 5′-A TTA CCG CGG CTG CTG G- 3′) [8,27]. Invitrogen Platinum PCR SuperMix High Fidelity was used for a PCR reaction of 35 cycles as follows- 94 °C for 30 s, 65 °C for 30 s, and 68 °C for 25 s. All PCR products were purified by size selection on 2% (wt/vol) agarose gel by electrophoresis. The concentration of purified DNA products was measured with Qubit dsDNA HS Assay Kit (Life Technologies, Carlsbad, US). Later, all the samples were pooled in equal concentrations, and sequencing was performed according to the manufacturer's protocol with an Ion S5 sequencer for 500 flows, which executed an average read length of >220 bp. Sequencing was carried out up to such depths to ensure that plateaus are obtained in rarefaction curves.
2.7. Estimation of taxonomic diversity for network analysis
To exclude DNA contamination, 100% aligned Kitome reads were excluded from sample reads with short-read aligner Bowtie2 (v. 2.2.4.4) in default stagnant with end-to-end and very sensitive alignment mode [30]. With the help of Fastx-toolkit 0.0.14, sequences extracted after kitome removal were once more filtered with a sequence length cutoff of 100 bp and quality value of 20 (http://hannonlab.cshl.edu/fastx_toolkit). Using modules from UPARSE later reads were grouped into OTUs (Operational Taxonomic Units) with 97% similarity [31], and singletons were discarded. The taxonomic affiliation of each OTU's sequence was ascertained by RDP Classifier using an 80% confidence level (http://rdp.cme.msu.edu/classifier/classifier.jsp).
The networks were generated using the plugin CoNet (version 1.1.1.beta) for Cytoscape (version 3.9.1) with row_minocc minimum occurrence of 3 across all three conditions (pre-fog, fog, and post-fog) [32]. In addition, five methods were selected for ensemble inference: Pearson correlation, Spearman correlation, Mutual information, Bray-Curtis dissimilarity, and Kullback-Leibler dissimilarity. Thresholds were not set manually, and we requested for top 300 edges for each method and enabled the “Top to bottom” edge selection parameter. Then, permutation, bootstrapping, and restoration of networks were performed according to the tutorial mentioned in CoNet-2017. (http://msysbiology.com/microbialnetworks/conet_new.php). It is worth mentioning that data size might impact the network analysis results. In the present study, maximum change in the present data size was 40% in foggy condition, and we found that if there is 40% change in data size, variation in the network analysis results is about ±1%.
2.8. Taxonomic profiling and diversity analysis
The taxonomic profiling of 16S amplicon data of three conditions, pre-fog (n = 3), fog (n = 5), and post-fog (n = 4), was performed in Galaxy platform [33]. The quality check of the raw data was performed using fastQC (Version 0.11.9), and quality was trimmed using Trim Galore! (Version 0.6.7) [34,35]. Kraken2 (Version 2.1.1) was used for taxonomic assignment, and Silva (Created: 2022-02-02T162959Z, kmer-len = 35, minimizer-len = 31, minimizer-spaces = 6, load-factor = 0.7) was used as the reference database for mapping the reads [36]. The confidence score was set to 0.1 to classify more reads. The report file from kraken2 and metadata files were combined into a biom format file using the kraken-biom (version 1.0.1) package [37]. The biom files were further used to calculate the alpha and beta diversity using the phyloseq (version 1.38.0) package in R programming language [38]. The unidentified reads were filtered out. Shannon's diversity index (alpha diversity) of three conditions was computed using the plot_richness function in the phyloseq package, and Bray–Curtis distances and Principal Coordinate Analysis (PCoA) values (beta diversity) were calculated using ordinate function.
2.9. Statistical analysis
One-way ANOVA (Analysis of variance), ANOSIM (Analysis of similarities), and SIMPER (Similarity Percentage) were performed for all three atmospheric conditions and different target organs of pathogens with PAST software (version 4.03), and p-values of <0.05 were considered to be statistically significant. ANOSIM and SIMPER were performed with the Bray-Curtis distance measure in the present study.
3. Results and discussions
3.1. Meteorological synopsis
In pre-fog condition, RH is noticed to be maximum (reached up to 96 %) with minimum temperature (up to 7 °C), developing a cold, wet atmosphere. Such an atmosphere is favorable for enhancement of water droplets into the air, commonly known as fog droplets, which can absorb the water-soluble aerosols in air [26,39]. Low wind speed supported to accumulate them over the measurement site with decreasing visibility. Pre-fog condition occurred mainly in the early morning, so inclined solar radiation could not reach the ground. As a result, air temperature remained low, and such atmospheric conditions were maintained for longer periods, with plenty of floating tiny fog droplets in the air. Wind speed became zero during this period, which made atmosphere calm to retain water droplets in the air, resulting in lower visibility. This condition is identified as a dense foggy condition and selected as a foggy period for the present study. However, some droplets underwent coagulation and deposited from air, resulting in a progressive fall in RH, as seen in Fig. 2(a).
Further, the ambient temperature increased as the day progressed, and fog started disappearing. This period is identified as a post-fog condition. Post-fog condition had a minimum RH (45 %) and a maximum temperature (24 °C). The atmosphere in post-fog condition was warmer and drier. Such an atmosphere could not support fog droplets to survive for longer periods, and thereby, fog droplets started evaporating, which helped to enhance visibility and allowed more solar radiation reaching to the ground. In addition, wind speed increased in post-fog condition, and the movement of wind provided additional support to remove the fog droplets from the measurement site, accelerating the fog disappearing process.
PBLH plays a crucial role in fog formation and disappearing process. PBLH was noticed to be minimum during pre-fog condition, maintained same in foggy condition, and maximum in post-fog condition. Pre-fog condition had lowest PBLH, indicating less space for floating water droplets into the air, that is, less ventilation and favored fog formation. Low PBLH in foggy condition also helped to keep the fog layer near the ground. During post-fog condition, PBLH was increased significantly, which caused fog dissipation by both advection horizontally and convection vertically with higher ventilation.
Back-trajectories mainly originated from western IGP and traveled through major urban cities like Delhi, Kanpur, and Lucknow before reaching the sampling site, as shown in Fig. 3. A similar wind pattern was also observed, blowing from west to central IGP. Due to low wind speed, a slow movement of airmass was coming from west to east IGP, favoring fog formation. In addition, RH and temperature near the sampling site were in the range of 80–90 % and 10–15 °C, respectively, which also supported fog formation, as shown by contour lines in Fig. 3.
3.2. Richness and diversity of atmospheric bacteria
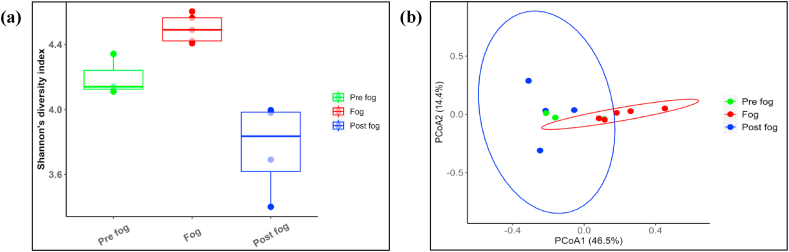
Shannon's diversity index was calculated from 12 amplicon sequencing datasets in pre-fog, fog, and post-fog conditions. Fig. 4(a) shows the boxes presenting the alpha diversity of each atmospheric condition, and each filled dot indicates one sample. The horizontal line within the boxes presents the median, and the vertical lines show the standard deviation of each condition. Shannon's diversity index variation is noticed from 3.4 to 4.6 over the measurement site. Shannon's diversity index varied from 4.4 to 4.6 in foggy condition, followed by a variation of 4.1–4.3 in pre-fog and 3.4 to 4.0 in post-fog condition, indicating fog as the most diverse condition with highest values of Shannon's diversity index.
Fig. 4.
(a) Boxes present Shannon's diversity index with standard deviations in pre-fog, fog and post fog conditions where each points present individual sample. (b) Beta diversity between three atmospheric conditions is represented as PCoA plot, where ellipse is observed for fog and post-fog conditions.
Beta diversity was also computed to understand the bacterial diversity in different atmospheric conditions, as shown in Fig. 4(b). Ellipses are formed depending on Bray-Curtis dissimilarity distances, helping to cluster the sampling points, as shown in the figure. Only two distinct ellipses can be formed representing fog and post-fog conditions, whereas an ellipse could not be formed due to insufficient samples (3) in pre-fog condition. It is worth mentioning here that the identification of pre-fog condition was challenging to recognize at the time of on-field sample collections, and thereby, many samples could not be included in the present study due to continuation of sample collection during pre-fog and fog conditions or fog and post-fog conditions. The fog samples exhibit a distinct diversity from the pre-fog and post-fog conditions, although there is a slight overlapping of two ellipses. The diversity of pre-fog and post-fog conditions is found to be quite similar.
3.3. Relation of airborne bacteria within networks in different atmospheric conditions
Simulation of networks helps to understand the possible interactions between airborne bacteria. To characterize the intricate web of relationships, topological features are considered in the present network analysis. The network features allow for rapid and easy comparisons between complicated data sets from different ecosystem types to investigate the essential characteristics of a specific habitat type influencing the diversity of microbial communities [40]. All genera present in the samples were used to construct networks individually in different atmospheric conditions. In the networks, each node represents bacterial genera, and edges represent possible significant interactions shown in [Fig. 5(a, b, c)], i.e., green is for copresence, and red is for mutual exclusion. Fig. 5 shows three individual networks in pre-fog, fog, and post-fog conditions. 81 nodes with 445 edges [copresence – 310 (70 %), mutual exclusion – 135 (30 %)] are present in a pre-fog network; 178 nodes with 618 edges [copresence – 398 (64 %), mutual exclusion – 220 (36 %)] are in foggy network, and 61 nodes with 331 edges [copresence – 282 (85 %), mutual exclusion – 49 (15 %)] are present in post-fog network. Lower copresence interactions are noticed in foggy condition due to the availability of nutrients within tiny liquid fog droplets that provides a suitable environment for bacterial growth, and ∼30% unique bacterial genera were present only in foggy condition. Availability of nutrients during foggy condition helped to enrich bacterial genera that might have been mutually excluded during pre-fog and post-fog conditions. In addition, poor visibility protects them from harmful solar radiation. However, higher copresence interactions are found in pre-fog and post-fog conditions due to a relatively less abundances in less suitable environmental conditions, thereby, comparatively more interactive with neighbours. Mutual exclusion of bacterial genera during pre-fog and post-fog conditions is relatively less dominant than in foggy condition, likely because of the elimination of bacteria from the population.
Fig. 5.
Network analysis showing interactions among bacteria (green edges-copresence, red edges-mutual exclusion) in (a) pre-fog, (b) fog, and (c) post-fog.
Rubellimicrobium (15 edges), Thioalkalivibrio (15 edges), and Methylocystis (14 edges) are the key genera in pre-fog condition. Methylocystis (21 edges) is also a key genus in foggy condition, followed by Cutibacterium (21 edges) and Azospira (19 edges). During post-fog condition, the key genera present are Agromyces (20 edges), Cellvibrio (18 edges), and Turicibacter (17 edges). Eliminating these key genera would lead to significant changes in the community [41]. Present investigation on network analysis indicates a unique bacterial ecosystem evolved in foggy condition over central IGP. A brief overview of these key genera is mentioned in Table 2.
Table 2.
Key genera in pre-fog, fog, and post-fog networks.
| Genera (Atmospheric Condition) | Isolation source | Temperature (°C) | pH | Specialty | References |
|---|---|---|---|---|---|
| Rubellimicrobium (Pre-fog) | Soil, air, and paper machines | 20-37 (Optimum 28) | 6.5–12.0 (Optimum 7) | – | [[45], [46], [47]] |
| Thioalkalivibrio (Pre-fog) | Soda lakes | 40-41 (Optimum 35) | 9.5–11.0 (Optimum 10) | Sulfur-oxidizing bacteria that survive in double extreme habitats with maximum salt concentration and alkaline pH. | [48] |
| Methylocystis (Pre-fog, Fog) | Boreal forest soil, uncontaminated ground water, coal mine drainage water, soil, raw water, lake sediment, marsh mud, and creek mud | 5-40 (Optimum 30–35) | 4.5–10.0 (Optimum 8.5–9.0) | Methylocystis are type II methanotrophs, which utilize methane and its derivatives as carbon source through serine pathway and executes essential environmental functions as they are a link in the global carbon cycle, act as nitrogen fixers, and can degrade a variety of organic contaminants. | [[49], [50], [51]] |
| Cutibacterium (Fog) | Skin microbiota, sebaceous areas of the face like glabella, alar crease, external auditory canal, and the back. | 31.8–36.6 | 4.2–7.9 | It is an opportunistic pathogen that causes breast infection, skin abscesses, infective endocarditis, and device-related infections in humans. | [14,15,[52], [53], [54], [55]] |
| Azospira (Fog) | Groundwater, plant roots, and grasses, especially in the roots of wild and cultured rice | 20-40 (Optimum 37) | 7 | – | [[56], [57], [58]] |
| Agromyces (Post-fog) | Soils of both saline and alkaline area, sea sediments, plant rhizosphere, and polluted sites | 10-37 (Optimum 28) | 6.0–10.0 (Optimum 7.0–8.0) | Pathogenic in nature and causes bacteremia in humans | [[59], [60], [61]] |
| Cellvibrio (Post-fog) | Soil, spring water, plant root-associated habitats, and gut of grass-feeding snails. | 10-35 (Optimum 25) | 6.5–9.5 | Cellvibrio hydrolyses cellulose and dextrin. Cellulases are gaining popularity these days due to their use in bioenergy and biofuel production. | [[62], [63], [64], [65]] |
| Turicibacter (Post-fog) | Gut of humans and animals, chicken eggshells, swine ileum, and soil and water. | 30-45 (Optimum 42) | 6.5–8.5 (Optimum 7.5) | – | [66,67] |
3.4. Pathogenic implications of fog-induced bacteria
A large number of unique bacterial genera are enriched in the occurrences of fog due to favorable environments like low temperatures with higher abundances of nutrients, water present in the floating fog droplets, protected from harmful UV rays by dimming down the incoming solar radiation in low visibility condition [[6], [7], [8]]. The pH of deposited fog water collected during sampling period is about 6.6 ± 0.1, which is suitable for the growth of most of the airborne bacteria, as mentioned in Table 2. In the present study, all the pathogenic bacterial genera present over measurement site are categorized into three groups depending on their targeted organs of humans viz, respiratory, skin, and oral. The total concentration of pathogenic genera during pre-fog, foggy, and post-fog conditions was 27, 42, and 29, respectively.
Skin infectious bacteria (66 %) are found to be dominating in pre-fog and foggy conditions, followed by respiratory (30 %) and oral (4 %) infectious bacteria. An increase in average OTU count is also evident for respiratory (44 %), skin (60 %), and oral (125 %) infectious bacteria during foggy condition, as shown in Fig. 6. A decrease of 85 % in respiratory and skin infectious bacteria is noticed in post-fog condition. However, bacteria causing oral infections are increased by 233 % in post-fog condition compared to foggy condition, which could be a slower growth rate of those bacteria.
Fig. 6.
Average abundances of air-borne pathogenic bacteria targeting respiratory, skin and oral organs for human being in pre-fog, fog, and post-fog atmospheric conditions. The vertical lines present standard deviations. Two Asterisks (**) indicate significance level of p < 0.01.
A rigorous statistical analysis was carried out in the current investigation. One-way ANOVA test shows a significant variation in total pathogenic potential from pre-fog to fog (F = 13.71, p < 0.01), from fog to post-fog (F = 15.74, p < 0.01), and overall (F = 21.78, p < 0.01) (Fig. 6). ANOSIM was also performed to obtain dissimilarity and skin infectious bacteria genera exhibited significant dissimilarity (p < 0.01); however, no significant results were noticed for respiratory and oral infectious bacteria. SIMPER was performed to quantify the overall dissimilarity in different atmospheric conditions. For respiratory, skin, and oral infectious bacteria, overall dissimilarity percentage varies from 35 to 45 %, 40–60 %, and 70–80 %, respectively. Oral infectious bacteria are found to be most dissimilar because of their high abundance in post-fog condition, which could be due to the delayed growth effect.
A detailed one-way ANOVA was performed for individual major pathogenic bacteria in different atmospheric conditions, and significant results were obtained for Paenibacillus, Mycobacterium, Cutibacteirum, Lysinibacillus, Beijerinckia, Capnocytophaga, and Streptococcus as shown in Fig. 7. The concentration of total pathogenic airborne bacteria like Cutibacterium, Lysinibacillus, Mycobacterium, Herbaspirillum, Paenibacillus, Tsukamurella, and Williamsia are found to be increased by 160 times in foggy condition than those in other two conditions, while their total loading in those conditions is found to be identical. Noticeably, Mycobacterium and Paenibacillus have been increased by a factor of two and four, respectively, in foggy condition. Some pathogenic bacteria like Herbaspirillum, Tsukamurella, and Williamsia could not be detected in pre-fog and post-fog conditions. As a result, unique airborne bacterial diversity was observed in fog, distinct from the pre-fog and post-fog conditions, noticed in alpha and beta diversity analysis (Fig. 4).
Fig. 7.
Average abundances of individual pathogenic bacterial genera in different atmospheric conditions. The horizontal lines represent standard deviations. Single (*) and double Asterisk (**) indicate significance level of p < 0.05, and p < 0.01, respectively for pre-fog and fog (green), fog & post-fog (blue), and overall (pink).
Cutibacterium, Herbaspirillum, Paenibacillus, and Tsukamurella are opportunistic pathogenic bacteria in foggy atmospheres, mainly targeting various parts of respiratory organs. Among them, Paenibacillus and Herbaspirillum can cause cancers in favorable conditions [18,42]. Cutibacterium is found to be the most important bacteria in foggy condition because it is directly responsible for various infections, including breast infection, skin abscesses, infective endocarditis, and device-related infections [14,15], as well as inter-link with other pathogenic bacteria present in the network found in foggy condition (Fig. 5(b)) and also mentioned in Table 3. Interestingly, Cutibacterium interacts with twenty-one other bacterial genera with a copresence relation. Out of twenty-one interactions, five bacterial genera are identified to be pathogenic in nature, i.e., Rothia, Parvimonas, Sedimentibacter, Brevundimonas, targeting lungs, blood infection via a wound, UTI, skin infection, etc. In the network, Xanthomonas is also pathogenic but for plants (Table 3).
Table 3.
Fog-induced pathogenic bacteria present in the network.
| Fog-induced pathogenic bacteria | Target organ | Type of Interaction | Interaction with genus | Pathogenicity | References |
|---|---|---|---|---|---|
| Cutibacterium (21) | Skin | Copresence | Faecalibacterium | Non-pathogenic | – |
| Bosea | |||||
| Aggregatilinea | |||||
| Desulfallas | |||||
| Oscillibacter | |||||
| Azospira | |||||
| Meiothermus | |||||
| Georgenia | |||||
| Ferrovibrio | |||||
| Shimazuella | |||||
| Hydrogenispora | |||||
| Idiomarina | |||||
| Methylobacter | |||||
| Sporosarcina | |||||
| GpV | |||||
| Methylocystis | |||||
| Rothia | Bacteremia, endocarditis, meningitis, peritonitis, bone and joint infections, pneumonia, skin and soft tissue infection, endophthalmitis, and prosthetic device infection. | [[68], [69], [70], [71], [72], [73]] | |||
| Parvimonas | It is a conditional pathogen that can cause infections in the human oral cavity, wounds, and other areas, as well as sepsis. It causes prosthetic joint infection, Lung abscess, Septic arthritis, intra-abdominal abscesses, and bacteremia. | [[74], [75], [76], [77]] | |||
| Sedimentibacter | Bacteremia | [78] | |||
| Brevundimonas | In humans, Brevundimonas is an opportunistic pathogen able to cause a range of hospital-acquired infections such as bacteremia, eye infection, peritonitis, urinary tract infection, and skin and soft tissue infection. | [79] | |||
| Xanthomonas | Xanthomonas spp. encompasses a wide range of plant pathogens that use numerous virulence factors for pathogenicity and fitness in plant hosts. | [80] | |||
| Lysinibacillus (1) | Pulmonary | Mutual Exclusion | Cellvibrio | Non-pathogenic | – |
| Mycobacterium (8) | Pulmonary | Mutual Exclusion | Rummeliibacillus | Non-pathogenic | – |
| Brevilactibacter | |||||
| Dokdonella | |||||
| Microbacterium | |||||
| Aciditerrimonas | |||||
| Propioniciclava | |||||
| Streptococcus |
Humans-Streptococcus pneumonia is responsible for the highest number of pneumonia cases worldwide. Pyogenic infections involving the mucous membranes, tonsils, skin, and deeper tissues, including pharyngitis, pyoderma, cellulitis, necrotizing fasciitis, toxic streptococcal syndrome, scarlet fever, septicemia, pneumonia, meningitis, respiratory failure. Animals- Septicemia in poultry. Meningitis, pneumonia, polyserositis, and polyarthritis in pigs. Mastitis in cattle. Fish- Meningoencephalitis, septicemia in fish. |
[[81], [82], [83]] | |||
| Schaalia | Dental caries, deep carious lesions around teeth. | [84] |
Among the fog-increased bacteria, Mycobacterium causes tuberculosis, which is initiated by the deposition of Mycobacterium tuberculosis on the alveoli of the lungs from aerosol droplets [16,17]. It is worth mentioning here that aerosols and bacteria have increased significantly in foggy conditions [8,43], which might support an increasing risk of such fatal diseases. Mycobacterium interacts with eight other bacteria in the network but with mutual exclusion. Among them, Streptococcus and Schaalia are also pathogenic in nature. It is to be noted that Streptococcus is infectious to humans and various animals, including fish and poultry (Table 3).
Lysinibacillus (p < 0.05) and Williamsia are increased in foggy condition and are responsible for respiratory diseases [19,44]. The present study represents that the industrially polluted region is highly loaded with various pathogenic airborne bacteria like Mycobacterium (p < 0.05), Williamsia, and Paenibacillus (p < 0.05) commonly isolated from multiple environments threatening human beings [18,20]. It is worth mentioning here that the sampling site is a rural area located at central IGP, fallen on the pathway of long-range transport of winter-time haze, as found from back-trajectory analysis. The industrial bacteria have increased over central IGP due to the movement of haze from nearby industrial regions traveling from west to east over IGP.
4. Summary and conclusions
Airborne samples were collected round the clock at a 4-h duration from 1 to January 14, 2021 at Arthauli (25.95°N, 85.10°E), a rural site located within the Vaishali district in the state of Bihar, over central IGP. Based on occurrences of fog over sampling site, the collected airborne samples were identified into three groups, i.e., pre-fog, fog, and post-fog conditions, and airborne bacteria were detected using NGS. The current study presents a distinct diversity of bacterial populations with a distinguishable complex network of a unique ecosystem in foggy condition. Such atmospheric condition is also enriched by several pathogenic bacterial genera, with a few opportunistic pathogens that target respiratory, skin, and oral organs in human.
The major conclusions are as follows.
-
i.
Distinct bacterial diversity has been identified in foggy condition than in pre- and post-fog conditions over central IGP due to unique types of bacteria detected in foggy condition. The reason could be the availability of water and nutrients in dimmed solar radiation condition, supporting their survival in a foggy atmosphere.
-
ii.
The present study identifies a complex network system of airborne bacteria in fog. In the foggy network, it has been noticed that maximum interaction occurred by an opportunistic pathogen, Cutibacterium, interacting with 21 other bacterial genera with statistically significant copresence relation. Interestingly, five of these interacting bacteria are also pathogenic for various infections like bacteremia, oral, respiratory, UTIs, etc.
-
iii.
A 40–60 % increase has been noticed in OTU count for respiratory and skin infections in foggy condition, while oral infections bacteria increased to 125 % and 233 % in fog and post-fog conditions, respectively.
-
iv.
Cutibacterium, Herbaspirillum, Paenibacillus, and Tsukamurella are opportunistic pathogenic bacteria enriched in foggy atmosphere and responsible for many diseases in various parts of respiratory organs.
-
v.
Most of the pathogenic bacteria enriched in foggy condition come from industrial and urban areas located in western direction to the measurement site of central IGP. Among them, Mycobacterium, Williamsia, Lysinibacillus, and Beijerinckia cause various respiratory and skin infections in humans.
-
vi.
Paenibacillus is an opportunist airborne bacterium enriched by four times noticed in foggy condition responsible for various health issues like chronic kidney disease, sickle cell disease, skin cancer, acute lymphoblastic leukemia, etc.
Present investigation indicates that exposure of humans to fog over central IGP for a longer period could harm their health, and more rigorous and regular investigations are required.
Data availability
The sequence files were deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI), USA, under the BioProject accession numbers PRJNA864626.
CRediT authorship contribution statement
Shahina Raushan Saikh: Writing – original draft, Software, Methodology, Investigation, Formal analysis, Data curation. Md Abu Mushtaque: Investigation. Antara Pramanick: Investigation. Jashvant Kumar Prasad: Investigation. Dibakar Roy: Software. Sudipto Saha: Validation, Supervision. Sanat Kumar Das: Writing – review & editing, Visualization, Supervision, Resources, Project administration, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Sanat Kumar Das reports financial support was provided by Science and Engineering Research Board. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This study was supported by the Bose Institute (Institutional Intramural Fund) and Science and Engineering Research Board (SERB), Government of India (GoI) (SERB grant no. CRG/2021/000619) (Extramural Fund). SRS, MAM, AP, JKP, and DR thank University Grants Commission (UGC), GoI for providing fellowship. We acknowledge the National Oceanic and Atmospheric Administration (NOAA), Air Resources Laboratory (ARL) for providing the HySPLIT model, National Centre for Medium Range Weather Forecasting (NCMRWF), Ministry of Earth Sciences (MoES), GoI for providing meteorological data. We also thank National Centers for Environmental Prediction-National Center for Atmospheric Research (NCEP-NCAR) for the spatial distribution of wind vectors and Meteorological and Oceanographic Satellite Data Archival Centre (MOSDAC), India Meteorological Department (IMD), GoI for providing fog intensity image from satellite observations.
References
- 1.Jaenicke R. Abundance of cellular material and proteins in the atmosphere. Science. 2005;308 doi: 10.1126/science.1106335. 73–73. [DOI] [PubMed] [Google Scholar]
- 2.Salem H., Gardner D.E. In: Atmospheric Microbial Aerosols. Lighthart B., Mohr A.J., editors. Springer US; Boston, MA: 1994. Health aspects of bioaerosols; pp. 304–330. [DOI] [Google Scholar]
- 3.Griffin D.W. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin. Microbiol. Rev. 2007;20:459–477. doi: 10.1128/CMR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Federici E., Petroselli C., Montalbani E., Casagrande C., Ceci E., Moroni B., La Porta G., Castellini S., Selvaggi R., Sebastiani B., Crocchianti S., Gandolfi I., Franzetti A., Cappelletti D. Airborne bacteria and persistent organic pollutants associated with an intense Saharan dust event in the Central Mediterranean. Sci. Total Environ. 2018;645:401–410. doi: 10.1016/j.scitotenv.2018.07.128. [DOI] [PubMed] [Google Scholar]
- 5.Kobziar L.N., Vuono D., Moore R., Christner B.C., Dean T., Betancourt D., Watts A.C., Aurell J., Gullett B. Wildland fire smoke alters the composition, diversity, and potential atmospheric function of microbial life in the aerobiome. ISME Commun. 2022;2:8. doi: 10.1038/s43705-022-00089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuzzi S., Mandrioli P., Perfetto A. Fog droplets—an atmospheric source of secondary biological aerosol particles. Atmos. Environ. 1997;31:287–290. doi: 10.1016/1352-2310(96)00160-4. [DOI] [Google Scholar]
- 7.Dong L., Qi J., Shao C., Zhong X., Gao D., Cao W., Gao J., Bai R., Long G., Chu C. Concentration and size distribution of total airborne microbes in hazy and foggy weather. Sci. Total Environ. 2016;541:1011–1018. doi: 10.1016/j.scitotenv.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Saikh S.R., Das S.K. Fog-induced alteration in airborne microbial community: a study over central Indo-Gangetic Plain in India. Appl. Environ. Microbiol. 2023;89 doi: 10.1128/aem.01367-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dueker M.E., O'Mullan G.D., Weathers K.C., Juhl A.R., Uriarte M. Coupling of fog and marine microbial content in the near-shore coastal environment. Biogeosciences. 2012;9:803–813. doi: 10.5194/bg-9-803-2012. [DOI] [Google Scholar]
- 10.Evans S.E., Dueker M.E., Logan J.R., Weathers K.C. The biology of fog: results from coastal Maine and Namib Desert reveal common drivers of fog microbial composition. Sci. Total Environ. 2019;647:1547–1556. doi: 10.1016/j.scitotenv.2018.08.045. [DOI] [PubMed] [Google Scholar]
- 11.Spencer R.C. Bacillus anthracis. J. Clin. Pathol. 2003;56:182–187. doi: 10.1136/jcp.56.3.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devanga Ragupathi N.K., Veeraraghavan B. Accurate identification and epidemiological characterization of Burkholderia cepacia complex: an update. Ann. Clin. Microbiol. Antimicrob. 2019;18:7. doi: 10.1186/s12941-019-0306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M.A., Rajandream M.-A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J.E., Taylor K., Whitehead S., Barrell B.G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;396 doi: 10.1038/24206. 190–190. [DOI] [PubMed] [Google Scholar]
- 14.Corvec S. Clinical and biological features of Cutibacterium (formerly propionibacterium) avidum , an underrecognized microorganism. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayslich C., Grange P.A., Dupin N. Cutibacterium acnes as an opportunistic pathogen: an update of its virulence-associated factors. Microorganisms. 2021;9:303. doi: 10.3390/microorganisms9020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003;16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jabir R.A., Rukmana A., Saleh I., Kurniawati T. In: Mycobacterium - Research and Development. Ribón W., editor. InTech; 2018. The existence of Mycobacterium tuberculosis in microenvironment of bone. [DOI] [Google Scholar]
- 18.Grady E.N., MacDonald J., Liu L., Richman A., Yuan Z.-C. Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Factories. 2016;15:203. doi: 10.1186/s12934-016-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.del Mar Tomas M., Moure R., Nieto J.A.S., Fojon S., Fernandez A., Diaz M., Villanueva R., Bou G. Williamsia muralis pulmonary infection. Emerg. Infect. Dis. 2005;11:1324–1325. doi: 10.3201/eid1108.050439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yassin A.F., Hupfer H. Williamsia deligens sp. nov., isolated from human blood. Int. J. Syst. Evol. Microbiol. 2006;56:193–197. doi: 10.1099/ijs.0.63856-0. [DOI] [PubMed] [Google Scholar]
- 21.Murray R.J., Aravena-Román M., Kämpfer P. Endophthalmitis due to Williamsia muralis. J. Med. Microbiol. 2007;56:1410–1412. doi: 10.1099/jmm.0.47270-0. [DOI] [PubMed] [Google Scholar]
- 22.Yassin A.F., Lombardi S.J., Fortunato S.J., McNabb P.C., Carr M.B., Trabue C.H. Perinatal sepsis caused by Williamsia serinedens infection in a 31-year-old pregnant woman. J. Clin. Microbiol. 2010;48:2626–2629. doi: 10.1128/JCM.00538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh V.P., Gupta T., Tripathi S.N., Jariwala C., Das U. Experimental study of the effects of environmental and fog condensation nuclei parameters on the rate of fog formation and dissipation using a new laboratory scale fog generation facility. Aerosol Air Qual. Res. 2011;11:140–154. doi: 10.4209/aaqr.2010.08.0071. [DOI] [Google Scholar]
- 24.Sawaisarje G.K., Khare P., Shirke C.Y., Deepakumar S., Narkhede N.M. Study of winter fog over Indian subcontinent : climatological perspectives. Mausam. 2014;65:19–28. doi: 10.54302/mausam.v65i1.858. [DOI] [Google Scholar]
- 25.Ghude S.D., Bhat G.S., Prabhakaran T., Jenamani R.K., Chate D.M., Safai P.D., Karipot A.K., Konwar M., Pithani P., Sinha V., Rao P.S.P., Dixit S.A., Tiwari S., Todekar K., Varpe S., Srivastava A.K., Bisht D.S., Murugavel P., Ali K., Mina U., Dharua M., Rao J., Padmakumari B., Hazra A., Nigam N., Shende U., Lal D.M., Chandra B.P., Mishra A.K., Kumar A., Hakkim H., Pawar H., Acharja P., Kulkarni R., Subharthi C., Balaji B., Varghese M., Bera S., Rajeevan M. Winter fog experiment over the Indo-Gangetic plains of India. Curr. Sci. 2017;112:767. doi: 10.18520/cs/v112/i04/767-784. [DOI] [Google Scholar]
- 26.Lubin D. Longwave radiative forcing of Indian Ocean tropospheric aerosol. J. Geophys. Res. 2002;107:8004. doi: 10.1029/2001JD001183. [DOI] [Google Scholar]
- 27.Roy C., Rameez M.J., Haldar P.K., Peketi A., Mondal N., Bakshi U., Mapder T., Pyne P., Fernandes S., Bhattacharya S., Roy R., Mandal S., O'Neill W.K., Mazumdar A., Mukhopadhyay S.K., Mukherjee A., Chakraborty R., Hallsworth J.E., Ghosh W. Microbiome and ecology of a hot spring-microbialite system on the Trans-Himalayan Plateau. Sci. Rep. 2020;10:5917. doi: 10.1038/s41598-020-62797-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stinson L.F., Keelan J.A., Payne M.S. Identification and removal of contaminating microbial DNA from PCR reagents: impact on low-biomass microbiome analyses. Lett. Appl. Microbiol. 2019;68:2–8. doi: 10.1111/lam.13091. [DOI] [PubMed] [Google Scholar]
- 29.Zubiria-Barrera C., Stock M., Neubert R., Vester A., Kulle A., Schneegans A., Leistner R., Gastmeier P., Slevogt H., Klassert T.E. A simple sequence-based filtering method for the removal of contaminants in low-biomass 16S rRNA amplicon sequencing approaches. J. Microbiol. Methods. 2020;178 doi: 10.1016/j.mimet.2020.106060. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 32.Faust K., Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Res. 2016;5:1519. doi: 10.12688/f1000research.9050.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Galaxy Community E. Afgan, Nekrutenko A., Grüning B.A., Blankenberg D., Goecks J., Schatz M.C., Ostrovsky A.E., Mahmoud A., Lonie A.J., Syme A., Fouilloux A., Bretaudeau A., Nekrutenko A., Kumar A., Eschenlauer A.C., DeSanto A.D., Guerler A., Serrano-Solano B., Batut B., Grüning B.A., Langhorst B.W., Carr B., Raubenolt B.A., Hyde C.J., Bromhead C.J., Barnett C.B., Royaux C., Gallardo C., Blankenberg D., Fornika D.J., Baker D., Bouvier D., Clements D., de Lima Morais D.A., Tabernero D.L., Lariviere D., Nasr E., Afgan E., Zambelli F., Heyl F., Psomopoulos F., Coppens F., Price G.R., Cuccuru G., Corguillé G.L., Von Kuster G., Akbulut G.G., Rasche H., Hotz H.-R., Eguinoa I., Makunin I., Ranawaka I.J., Taylor J.P., Joshi J., Hillman-Jackson J., Goecks J., Chilton J.M., Kamali K., Suderman K., Poterlowicz K., Yvan L.B., Lopez-Delisle L., Sargent L., Bassetti M.E., Tangaro M.A., van den Beek M., Čech M., Bernt M., Fahrner M., Tekman M., Föll M.C., Schatz M.C., Crusoe M.R., Roncoroni M., Kucher N., Coraor N., Stoler N., Rhodes N., Soranzo N., Pinter N., Goonasekera N.A., Moreno P.A., Videm P., Melanie P., Mandreoli P., Jagtap P.D., Gu Q., Weber R.J.M., Lazarus R., Vorderman R.H.P., Hiltemann S., Golitsynskiy S., Garg S., Bray S.A., Gladman S.L., Leo S., Mehta S.P., Griffin T.J., Jalili V., Yves V., Wen V., Nagampalli V.K., Bacon W.A., de Koning W., Maier W., Briggs P.J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022;50:W345–W351. doi: 10.1093/nar/gkac247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrews S. Babraham Institute; Cambridge, United Kingdom: 2010. FastQC: a Quality Control Tool for High Throughput Sequence Data, Babraham Bioinformatics. [Google Scholar]
- 35.Krueger F., James F., Ewels P., Afyounian E., Schuster-Boeckler B. 2021. FelixKrueger/TrimGalore: v0.6.7 - DOI via Zenodo. [DOI] [Google Scholar]
- 36.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dabdoub S. 2016. Kraken-Biom: Enabling Interoperative Format Conversion for Kraken Results.https://github.com/smdabdoub/kraken-biom [Google Scholar]
- 38.McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das S.K., Jayaraman A., Misra A. Fog-induced variations in aerosol optical and physical properties over the Indo-Gangetic Basin and impact to aerosol radiative forcing. Ann. Geophys. 2008;26:1345–1354. doi: 10.5194/angeo-26-1345-2008. [DOI] [Google Scholar]
- 40.Barberán A., Henley J., Fierer N., Casamayor E.O. Structure, inter-annual recurrence, and global-scale connectivity of airborne microbial communities. Sci. Total Environ. 2014;487:187–195. doi: 10.1016/j.scitotenv.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 41.Steele J.A., Countway P.D., Xia L., Vigil P.D., Beman J.M., Kim D.Y., Chow C.-E.T., Sachdeva R., Jones A.C., Schwalbach M.S., Rose J.M., Hewson I., Patel A., Sun F., Caron D.A., Fuhrman J.A. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011;5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhital R., Paudel A., Bohra N., Shin A.K. Herbaspirillum infection in humans: a case report and review of literature. Case Rep. Infect. Diseases. 2020;2020:1–6. doi: 10.1155/2020/9545243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das S.K., Chatterjee A., Ghosh S.K., Raha S. Fog-induced changes in optical and physical properties of transported aerosols over sundarban, India. Aerosol Air Qual. Res. 2015;15:1201–1212. doi: 10.4209/aaqr.2014.11.0287. [DOI] [Google Scholar]
- 44.Sulaiman I.M., Hsieh Y.-H., Jacobs E., Miranda N., Simpson S., Kerdahi K. Identification of Lysinibacillus fusiformis isolated from cosmetic samples using MALDI-TOF MS and 16S rRNA sequencing methods. J. AOAC Int. 2018;101:1757–1762. doi: 10.5740/jaoacint.18-0092. [DOI] [PubMed] [Google Scholar]
- 45.Denner E.B.M., Kolari M., Hoornstra D., Tsitko I., Kämpfer P., Busse H.-J., Salkinoja-Salonen M. Rubellimicrobium thermophilum gen. nov., sp. nov., a red-pigmented, moderately thermophilic bacterium isolated from coloured slime deposits in paper machines. Int. J. Syst. Evol. Microbiol. 2006;56:1355–1362. doi: 10.1099/ijs.0.63751-0. [DOI] [PubMed] [Google Scholar]
- 46.Dastager S.G., Lee J.-C., Ju Y.-J., Park D.-J., Kim C.-J. Rubellimicrobium mesophilum sp. nov., a mesophilic, pigmented bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2008;58:1797–1800. doi: 10.1099/ijs.0.65590-0. [DOI] [PubMed] [Google Scholar]
- 47.Jiang L.-Q., Zhang K., Li G.-D., Wang X.-Y., Shi S.-B., Li Q.-Y., An D.-F., Lang L., Wang L.-S., Jiang C.-L., Jiang Y. Rubellimicrobium rubrum sp. nov., a novel bright reddish bacterium isolated from a lichen sample. Antonie Leeuwenhoek. 2019;112:1739–1745. doi: 10.1007/s10482-019-01304-5. [DOI] [PubMed] [Google Scholar]
- 48.Ahn A.-C., Meier-Kolthoff J.P., Overmars L., Richter M., Woyke T., Sorokin D.Y., Muyzer G. Genomic diversity within the haloalkaliphilic genus Thioalkalivibrio. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gulledge J., Ahmad A., Steudler P.A., Pomerantz W.J., Cavanaugh C.M. Family- and genus-level 16S rRNA-targeted oligonucleotide probes for ecological studies of methanotrophic bacteria. Appl. Environ. Microbiol. 2001;67:4726–4733. doi: 10.1128/AEM.67.10.4726-4733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb H.K., Ng H.J., Ivanova E.P. In: The Prokaryotes. Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. The family methylocystaceae; pp. 341–347. [DOI] [Google Scholar]
- 51.Tikhonova E.N., Grouzdev D.S., Avtukh A.N., Kravchenko I.K. Methylocystis silviterrae sp.nov., a high-affinity methanotrophic bacterium isolated from the boreal forest soil. Int. J. Syst. Evol. Microbiol. 2021;71 doi: 10.1099/ijsem.0.005166. [DOI] [PubMed] [Google Scholar]
- 52.Wilson M. Cambridge University Press; New York: 2005. Microbial Inhabitants of Humans: Their Ecology and Role in Health and Disease. [Google Scholar]
- 53.Kong H.H., Segre J.A. Skin microbiome: looking back to move forward. J. Invest. Dermatol. 2012;132:933–939. doi: 10.1038/jid.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grice E.A. The intersection of microbiome and host at the skin interface: genomic- and metagenomic-based insights. Genome Res. 2015;25:1514–1520. doi: 10.1101/gr.191320.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fournière M., Latire T., Souak D., Feuilloley M.G.J., Bedoux G. Staphylococcus epidermidis and Cutibacterium acnes: two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms. 2020;8:1752. doi: 10.3390/microorganisms8111752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reinhold-Hurek B., Hurek T., Gillis M., Hoste B., Vancanneyt M., Kersters K., De Ley J. Azoarcus gen. Nov., nitrogen-fixing proteobacteria associated with roots of kallar grass (Leptochloa fusca (L.) Kunth), and description of two species, azoarcus indigens sp. nov. and azoarcus communis sp. nov. Int. J. Syst. Bacteriol. 1993;43:574–584. doi: 10.1099/00207713-43-3-574. [DOI] [Google Scholar]
- 57.Reinhold-Hurek B., Hurek T. In: The Genera Azoarcus, Azovibrio, Azospira and Azonexus. Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., editors. Springer; New York: 2006. pp. 873–891. (The Prokaryotes). New York, NY. [DOI] [Google Scholar]
- 58.Bae H.-S., Rash B.A., Rainey F.A., Nobre M.F., Tiago I., Da Costa M.S., Moe W.M. Description of Azospira restricta sp. nov., a nitrogen-fixing bacterium isolated from groundwater. Int. J. Syst. Evol. Microbiol. 2007;57:1521–1526. doi: 10.1099/ijs.0.64965-0. [DOI] [PubMed] [Google Scholar]
- 59.Hamada M., Shibata C., Tamura T., Suzuki K. Agromyces marinus sp. nov., a novel actinobacterium isolated from sea sediment. J. Antibiot. 2014;67:703–706. doi: 10.1038/ja.2014.60. [DOI] [PubMed] [Google Scholar]
- 60.Sridhar S., Wang A.Y.M., Chan J.F.W., Yip C.C.Y., Lau S.K.P., Woo P.C.Y., Yuen K.-Y. First report of human infection by agromycesmediolanus, a gram-positive organism found in soil. J. Clin. Microbiol. 2015;53:3377–3379. doi: 10.1128/JCM.01508-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evtushenko L.I., Ariskina E.V., Prisyazhnaya N.V., Starodumova I.P. In: Bergey's Manual of Systematics of Archaea and Bacteria. first ed. Whitman W.B., Rainey F., Kämpfer P., Trujillo M., Chun J., DeVos P., Hedlund B., Dedysh S., editors. Wiley; 2017. Agromyces; pp. 1–49. [DOI] [Google Scholar]
- 62.DeBoy R.T., Mongodin E.F., Fouts D.E., Tailford L.E., Khouri H., Emerson J.B., Mohamoud Y., Watkins K., Henrissat B., Gilbert H.J., Nelson K.E. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J. Bacteriol. 2008;190:5455–5463. doi: 10.1128/JB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ofek-Lalzar M., Sela N., Goldman-Voronov M., Green S.J., Hadar Y., Minz D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014;5:4950. doi: 10.1038/ncomms5950. [DOI] [PubMed] [Google Scholar]
- 64.Xie Z., Lin W., Luo J. Genome sequence of Cellvibrio pealriver PR1, a xylanolytic and agarolytic bacterium isolated from freshwater. J. Biotechnol. 2015;214:57–58. doi: 10.1016/j.jbiotec.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y., Xu J., Riera N., Jin T., Li J., Wang N. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome. 2017;5:97. doi: 10.1186/s40168-017-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maki J.J., Looft T. Turicibacter bilis sp. nov., a novel bacterium isolated from the chicken eggshell and swine ileum. Int. J. Syst. Evol. Microbiol. 2022;72 doi: 10.1099/ijsem.0.005153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lynch J.B., Gonzalez E.L., Choy K., Faull K.F., Jewell T., Arellano A., Liang J., Yu K.B., Paramo J., Hsiao E.Y. Turicibacter strains differentially modify bile acids and host lipids. Microbiology. 2022 doi: 10.1101/2022.06.27.497673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaccher S., Cordiali R., Osimani P., Manso E., De Benedictis F.M. Bacteremia caused by Rothia mucilaginosa in a patient with shwachman-diamond syndrome. Infection. 2007;35:209–210. doi: 10.1007/s15010-007-6284-8. [DOI] [PubMed] [Google Scholar]
- 69.Lee A.B., Harker-Murray P., Ferrieri P., Schleiss M.R., Tolar J. Bacterial meningitis from Rothia mucilaginosa in patients with malignancy or undergoing hematopoietic stem cell transplantation. Pediatr. Blood Cancer. 2008;50:673–676. doi: 10.1002/pbc.21286. [DOI] [PubMed] [Google Scholar]
- 70.Trivedi M.N., Malhotra P. Rothia prosthetic knee joint infection. J. Microbiol. Immunol. Infect. 2015;48:453–455. doi: 10.1016/j.jmii.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 71.Keng T.C., Ng K.P., Tan L.P., Chong Y.B., Wong C.M., Lim S.K. Rothia dentocariosa repeat and relapsing peritoneal dialysis-related peritonitis: a case report and literature review. Renal Failure. 2012;34:804–806. doi: 10.3109/0886022X.2012.678208. [DOI] [PubMed] [Google Scholar]
- 72.Bruminhent J., Tokarczyk M.J., Jungkind D., DeSimone J.A. Rothia mucilaginosa prosthetic device infections: a case of prosthetic valve endocarditis. J. Clin. Microbiol. 2013;51:1629–1632. doi: 10.1128/JCM.03173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cho E.-J., Sung H., Park S.-J., Kim M.-N., Lee S.-O. Rothia mucilaginosa pneumonia diagnosed by quantitative cultures and intracellular organisms of bronchoalveolar lavage in a lymphoma patient. Ann. Lab. Med. 2013;33:145–149. doi: 10.3343/alm.2013.33.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baghban A., Gupta S. Parvimonas micra: a rare cause of native joint septic arthritis. Anaerobe. 2016;39:26–27. doi: 10.1016/j.anaerobe.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 75.Shenoy P.A., Gupta R., Vishwanath S., Naik M.A., Shetty S., Chawla K. Parvimonas micra and fusobacterium nucleatum septic arthritis: a rare anaerobic double trouble. Asian J. Pharmaceut. Clin. Res. 2018;11:5. doi: 10.22159/ajpcr.2018.v11i12.28499. [DOI] [Google Scholar]
- 76.Huang Z., Zhang C., Li W., Fang X., Wang Q., Xing L., Li Y., Nie X., Yang B., Zhang W. Metagenomic next-generation sequencing contribution in identifying prosthetic joint infection due to Parvimonas micra: a case report. J. Bone Joint Infect. 2019;4:50–55. doi: 10.7150/jbji.30615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yun S.S., Cho H.S., Heo M., Jeong J.H., Lee H.R., Ju S., Kim J.-Y., You J.W., Cho Y.J., Jeong Y.Y., Kim H.C., Lee J.D., Lee S.J. Lung abscess by Actinomyces odontolyticus and Parvimonas micra co-infection presenting as acute respiratory failure: a case report. Medicine. 2019;98 doi: 10.1097/MD.0000000000016911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woo P.C.Y., Teng J.L.L., Leung K., Lau S.K.P., Wong M.K.M., Yuen K. Bacteremia in a patient with colonic carcinoma caused by a novel Sedimentibacter species: Sedimentibacter hongkongensis sp. nov. Diagn. Microbiol. Infect. Dis. 2004;50:81–87. doi: 10.1016/j.diagmicrobio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 79.Ryan M.P., Pembroke J.T. Brevundimonas spp: emerging global opportunistic pathogens. Virulence. 2018;9:480–493. doi: 10.1080/21505594.2017.1419116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Timilsina S., Potnis N., Newberry E.A., Liyanapathiranage P., Iruegas-Bocardo F., White F.F., Goss E.M., Jones J.B. Xanthomonas diversity, virulence and plant–pathogen interactions. Nat. Rev. Microbiol. 2020;18:415–427. doi: 10.1038/s41579-020-0361-8. [DOI] [PubMed] [Google Scholar]
- 81.Chanter N. Streptococci and enterococci as animal pathogens. Soc. Appl. Bacteriol. Symp. Ser. 1997;26:100S–109S. [PubMed] [Google Scholar]
- 82.Berridge B.R., Fuller J.D., De Azavedo J., Low D.E., Bercovier H., Frelier P.F. Development of specific nested oligonucleotide PCR primers for the Streptococcus iniae 16S-23S ribosomal DNA intergenic spacer. J. Clin. Microbiol. 1998;36:2778–2781. doi: 10.1128/JCM.36.9.2778-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cunningham M.W. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 2000;13:470–511. doi: 10.1128/CMR.13.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reimer L.C., Vetcininova A., Carbasse J.S., Söhngen C., Gleim D., Ebeling C., Overmann J. Bac Dive in 2019: bacterial phenotypic data for High-throughput biodiversity analysis. Nucleic Acids Res. 2019;47:D631–D636. doi: 10.1093/nar/gky879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence files were deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI), USA, under the BioProject accession numbers PRJNA864626.