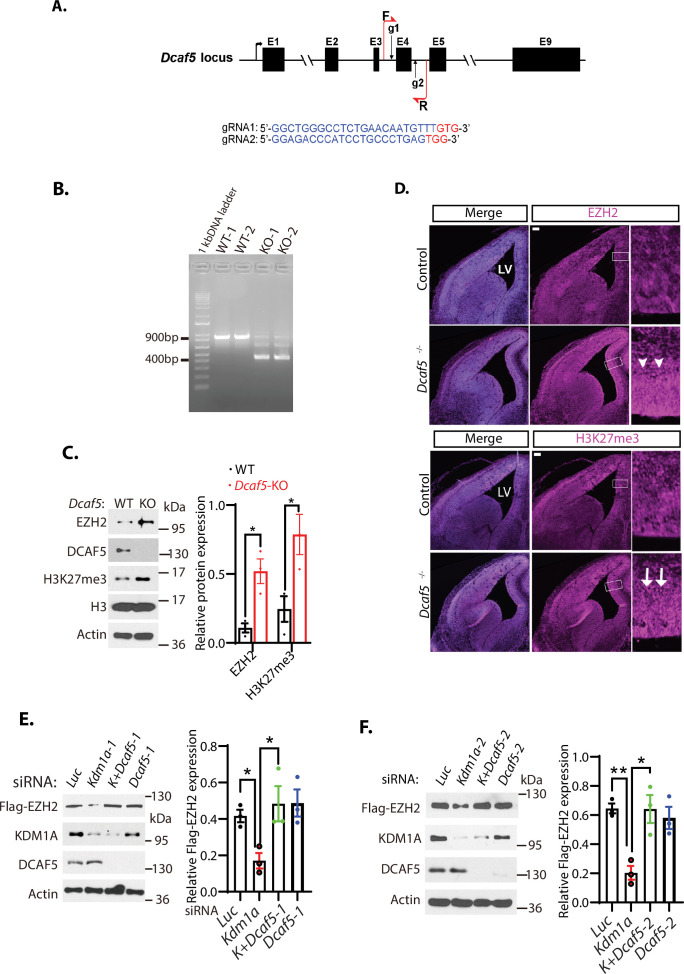
Figure 3. Loss of Dcaf5 stabilizes EZH2 protein.
(A) The strategy to delete the exon 4 of the mouse Dcaf5 gene by CRISPR-Cas9 gene edition with two guide RNAs (gRNAs). (B) Genome typing of the wild-type (WT) and Dcaf5 knock-out (KO) mice by PCR. (C) Western blot analysis of EZH2 and H3K27me3 proteins in the brains of the wild-type control and homozygous Dcaf5 deletion mice using the indicative antibodies. (D) Accumulation of EZH2 and H3K27me3 proteins in mouse Dcaf5 deleted embryonic brains. Immunostainings of anti-EZH2 and H3K27me3 in the coronal sections of the mouse embryonic brains of the wild-type control and Dcaf5 at E15.5. Scale bars, 100 μm. Arrows and arrowheads indicate the expression regions of EZH2 and H3K27me3, respectively. Boxed regions are enlarged on the right panels. LV: lateral ventricle. (E) Silencing of Dcaf5 re-stabilizes the protein levels of EZH2 in Kdm1a deficient cells. H1299 cells expressing stably expressed Flag-EZH2 were transfected with 50 nM siRNAs of luciferase (Luc), Kdm1a-1, Kdm1a and Dcaf5-1, and Dcaf5-1 siRNAs. The indicated proteins were analyzed by Western blotting. (F) Silencing of Dcaf5 re-stabilizes the protein levels of EZH2 in Kdm1a deficient cells. H1299 cells expressing stably expressed Flag-EZH2 were transfected with 50 nM siRNAs of luciferase (Luc), Kdm1a-2, Kdm1a-2+Dcaf5-2, and Dcaf5-2 siRNAs. The indicated proteins were analyzed by Western blotting. Band intensities in (C), (E), and (F) were quantified and normalized to that of the histone H3 or luciferase control. Significance was indicated as a two-tailed, unpaired, t-test. Values are expressed as the mean ± SEM. *p<0.05. **p<0.01.