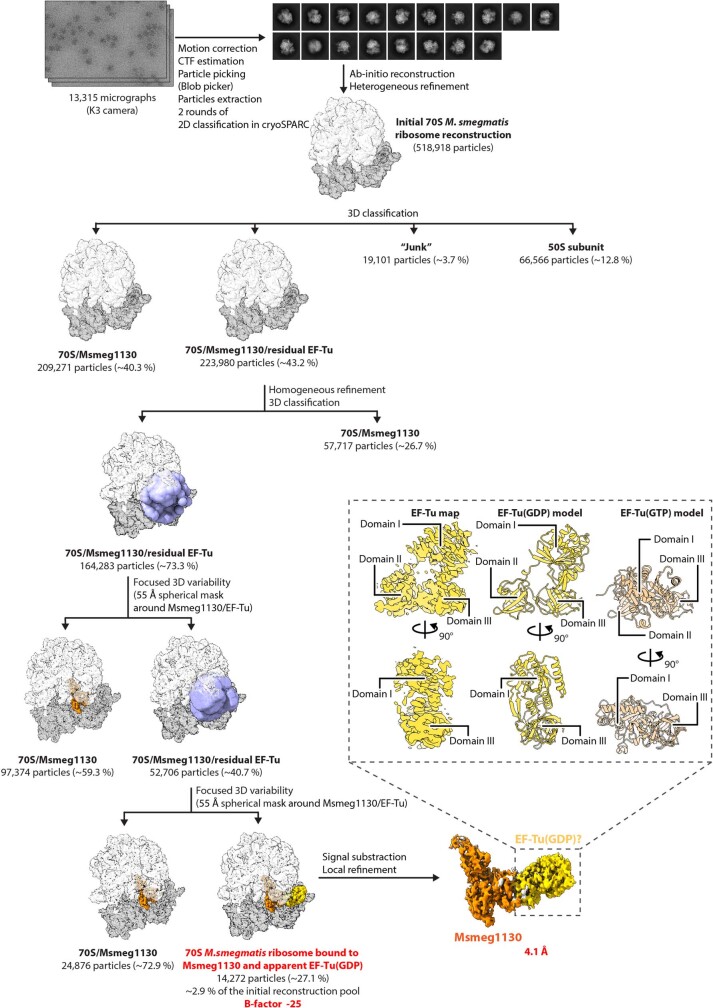
Extended Data Fig. 6. Cryo-EM data processing workflow using CryoSPARC for M. smegmatis ribosomes in complex with Msmeg1130 and EF-Tu in the presence of GDPCP.
The inset compares the EF-Tu signal with EF-Tu structures in the GTP and GDP states and shows that the EF-Tu density is not sufficient to build its molecular model but is sufficient to observe the overall conformation of the EF-Tu molecule. This conformation is consistent with the GDP-bound conformation of EF-Tu rather than the GTP-bound conformation, which likely stems from the residual amounts of GDP copurifying with our recombinant EF-Tu sample.