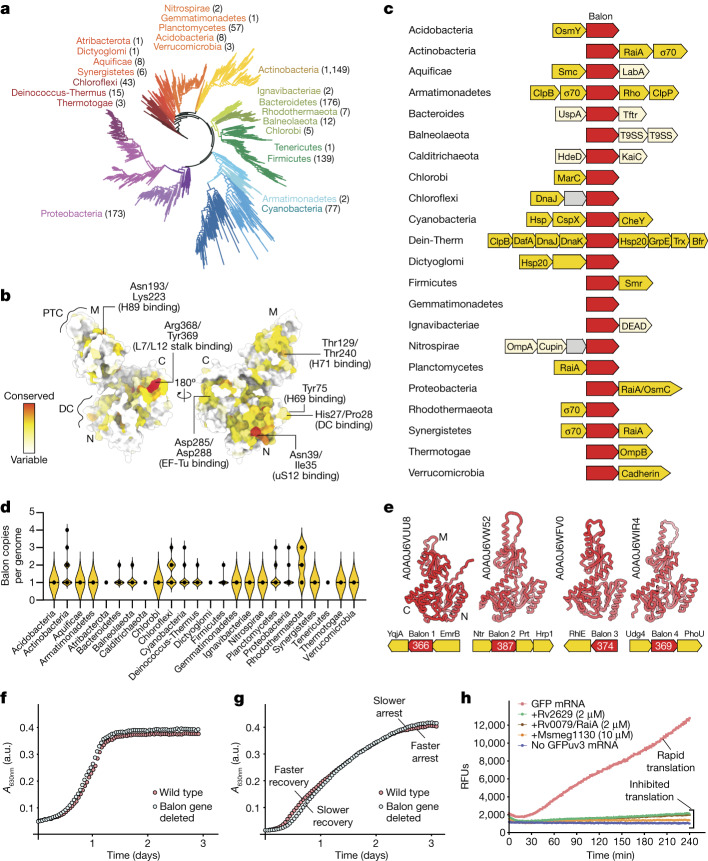
Fig. 2. Balon-coding genes are ubiquitous among bacteria and are often found in stress-response operons.
a, The bacterial tree of life shows that Balon homologues are found in most bacterial lineages, encompassing 1,572 representative bacteria from 23 different phyla. b, An atomic model of Balon (coloured by sequence conservation) illustrating high conservation of residues responsible for ribosome recognition. c, Operon schematics illustrating the genetic context of Balon-coding genes (red) in selected bacterial phyla. Balon-coding genes are typically found in operons that encode stress response factors (light orange). These include chaperones (Hsp20, DnaK and DnaJ), alternative σ70 factors, factors of acid tolerance (HdeD) and osmotic stress tolerance (OsmB and OsmY), ribosome hibernation (RaiA), ribosome repair (RtcB) and multidrug resistance (Smr, MarC and EmrB). Dein-Therm, Deinococcus-Thermus. d, Violin plots showing that in 603 species (38% of the analysed species), Balon homologues are encoded by two, three or four gene copies located in different genomic loci. e, As an example of a genome with several Balon orthologues, here we depict the four operons encoding Balon orthologues from Mycolicibacterium chubuense. Notably, one of these copies (Balon 1) resides in an operon with the multidrug export protein EmrB, and another copy (Balon 2) is located in an operon with hypoxia-response factors. Their predicted structures (AlphaFold) indicate a common core architecture. f, Growth curves of the wild-type and Balon-deficient strains of P. arcticus in a rich growth medium. g, Growth curves of the wild-type and Balon-deficient strains of P. arcticus during their recovery from the long-term stationary phase (3 months). h, Plot illustrating translation of the reporter protein GFP in the absence and presence of the hibernation factor RaiA (also known as Rv0079) or mycobacterial homologues of Balon—Rv2629 and Msmeg1130. RFUs, relative fluorescence units.