Fig. 4. Balon has a dissimilar mechanism to engage with the ribosomal active sites compared to aeRF1 and Pelota.
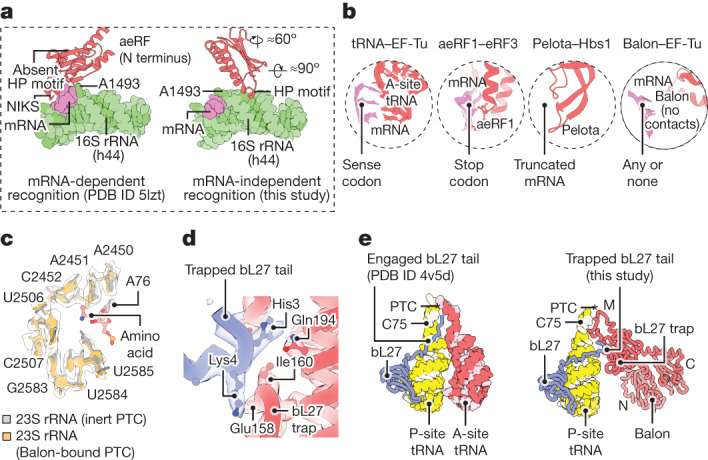
a,b, Zoomed-in views illustrating mRNA-independent recognition of the ribosome by Balon. a, Comparison illustrating the dissimilar mechanisms by which aeRF1 and Balon (red) recognize the decoding centre of the ribosome. 16S rRNA (green) and mRNA (pink) fragments are shown for simplicity. b, Comparison of decoding centre-binding strategies used by Balon and other ligands of the ribosomal A site. c–e, Zoomed-in views illustrating that Balon-bound ribosomes have an inactive conformation of the catalytic site. c, Superposed structures showing that the conformation of the peptidyl-transferase centre in Balon-bound ribosomes is identical to the inert state observed in ribosomes with a vacant A site and a peptidyl-tRNA-bound P site (Protein Data Bank (PDB) code 5mzd; rRNA backbone RMSD of 0.3 Å). d, A segment of the cryo-EM map showing bL27 protein (blue) bound to the bL27 trap of Balon (red) in the 70S ribosome (state 2). e, Comparison of the ribosomal protein bL27 (blue) in the structure of actively translating ribosomes (left) and ribosomes bound to Balon (right).
