Fig. 5. Balon participates in ribosome hibernation in complex with the GDP-bound form of EF-Tu.
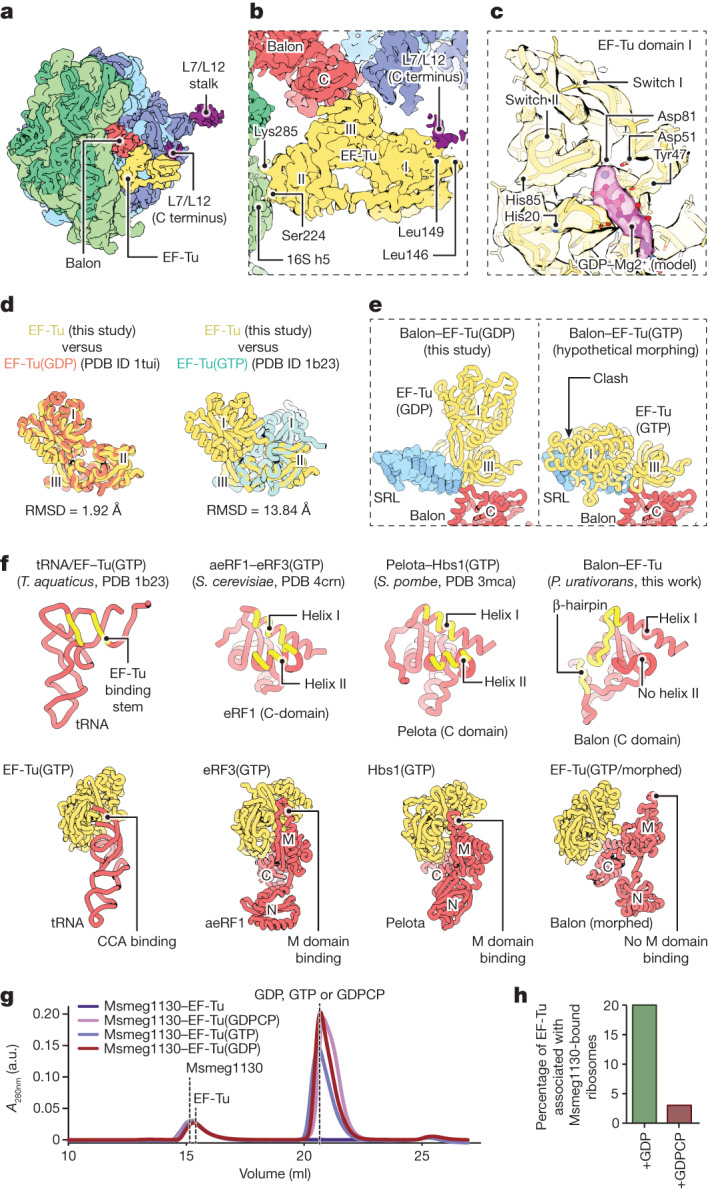
a, A cryo-EM map (filtered to 6 Å resolution) showing a P. urativorans ribosome bound to Balon and EF-Tu. b, A cryo-EM map focusing on the EF-Tu-binding sites illustrating that EF-Tu is attached to Balon-bound ribosomes through contacts with Balon, the C-terminal domain of the L7/L12 stalk (protein bL12) and the 16S rRNA helix h5. c, The structure of the EF-Tu nucleotide-binding domain and density consistent with the presence of GDP (the cryo-EM map is filtered to 4 Å resolution). d, A comparison of EF-Tu structures observed in this study and determined previously. EF-Tu molecules that are bound to Balon most closely resemble the GDP-bound conformation. e, Aligned structures showing that EF-Tu cannot adopt the GTP-bound conformation (observed in PDB code 1b23) while maintaining an interface with the C-terminal domain of Balon owing to a clash between EF-Tu domain I and the sarcin–ricin loop (SRL). f, Comparison of intramolecular interaction surfaces in four biological complexes, including EF-Tu–tRNA, eRF3–aeRF1, Hbs1–Pelota and EF-Tu–Balon. The upper panels highlight (in yellow) residues that recognize domain III of EF-Tu, or the EF-Tu homologues eRF3 and Hbs1. The lower panels compare complexes of EF-Tu(GTP)–tRNA, eRF3(GTP)–aeRF1 and Hbs1(GTP)–Pelota and the hypothetical complex of EF-Tu(GTP)–Balon, in which the Balon molecule is morphed to resemble the aeRF1 conformation in the eRF3(GTP)–aeRF1 complex. g, Size-exclusion chromatography shows that EF-Tu and Balon (Msmeg1130) do not form a stable complex even in the presence of GTP or GDPCP. h, Cryo-EM-based measurements of EF-Tu association with Msmeg1130-bound M. smegmatis ribosomes illustrate that EF-Tu effectively binds to Balon (Msmeg1130) in the ribosome only in the presence of GDP but not in the presence of GDPCP.
