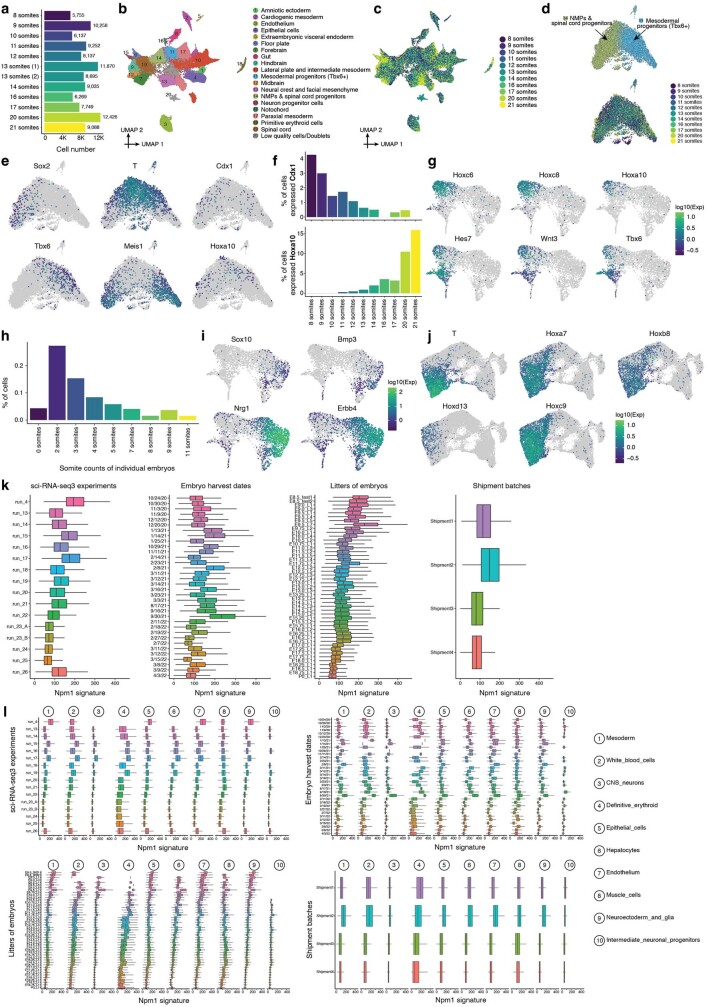
Extended Data Fig. 4. Transcriptional heterogeneity in the posterior embryo during early somitogenesis.
a, A validation sci-RNA-seq3 dataset of mouse embryos from somites 8 to 21. To validate findings related to differences between embryos staged with early vs. late somite counts, particularly in NMPs, we profiled another 12 precisely staged mouse embryos, ranging from 8 to 21 somites, in an independent sci-RNA-seq3 experiment. The resulting library was sequenced on an Illumina NextSeq 2000, resulting in 104,671 cells in total, with a median UMI count of 513 and a median gene count of 446 per cell. The number of cells profiled from each embryo. b, 2D UMAP visualization of the validation dataset (all cell types). c, The same UMAP as in panel b, with cells colored by somite count of the originating embryo. d, Re-embedded 2D UMAP of 9,686 cells from NMPs & spinal cord progenitors (cluster 11) and mesodermal progenitors (Tbx6 + ) (cluster 14) in panel b. Cells are colored by either the original annotation (top) or somite count (bottom). e, The same UMAP as in panel d, colored by gene expression of marker genes which appear specific to different subpopulations of NMPs: column 1: differences between neuroectodermal (Sox2 + ) vs. mesodermal (Tbx6 + ) fates; column 2: the differentiation of bipotential NMPs (T +, Meis1-) towards either fate; column 3: earlier (Cdx1 + ) vs. later (Hoxa10 + ) NMPs. References for marker genes are provided in Supplementary Table 12. f, Within the cells shown in panel d, the proportion of cells (y-axis) which express either Cdx1 (top) or Hoxa10 (bottom) are plotted as a function of somite count of the originating embryo. g, Transcriptional heterogeneity in the posterior embryo during the early somitogenesis. The same UMAP as in Fig. 2g, colored by gene expression of marker genes which appear specific to the subpopulation of notochord cluster that is Noto +, including posterior Hox genes (Hoxc6, Hoxc8, Hoxa10), and genes involved in Notch signaling (Hes7), Wnt signaling (Wnt3) and mesodermal differentiation (Tbx6). h, Cell proportions falling into the ciliated nodal cell cluster for embryos with different somite counts. i, The same UMAP as in Fig. 2g, colored by gene expression of marker genes which appear specific to the subpopulation of the notochord Noto- and more strongly Shh +, including Sox10, Bmp3, Nrg1, and Erbb4. j, The same UMAP as in Fig. 2i, colored by gene expression of marker genes which appear specific to the posterior gut endoderm, including T, Hoxa7, Hoxb8, Hoxd13, and Hoxc9. k, Checking the consistency of Npm1 signatures across different batches. First, we downsampled the dataset to ~1 M cells using geosketch79 and performed k-means clustering to ensure that each cluster contained roughly 500 cells. Second, we aggregated UMI counts for cells within each cluster to generate 2,289 meta-cells, and normalized the UMI counts for each meta-cell followed by log2-transformation. Third, we performed Pearson correlation between each protein-coding gene and Npm1, and selected genes with correlation coefficients > 0.6 (738 genes, ~3% of the total protein coding genes). A gene set enrichment analysis suggests that the module is associated with RNP complexes (corrected p-value = 1.4e-105), cytoplasmic translation (corrected p-value = 2.8e-90), and ribosomal proteins (corrected p-value = 7.4e-71). Finally, we summed the normalized UMI counts of these genes to calculate a Npm1 signature for individual cells. The resulting Npm1 signatures are subsetted in four plots, from left to right: by sci-RNA-seq3 experiment, embryo harvest date, litter of embryos, or shipment batch. l, Same as panel k, but further stratified by the top 10 abundant major cell clusters. Boxplots, in panel k (n = 1,144,141 cells) and l (n = 299,725 cells in Mesoderm, n = 127,150 cells in White blood cells, n = 104,205 cells in CNS neurons, n = 73,005 cells in Definitive erythroid, n = 66,772 cells in Epithelial cells, n = 64,845 cells in Hepatocytes, n = 62,951 cells in Endothelium, n = 61,249 cells in Muscle cells, n = 52,748 cells in Neuroectoderm and glia, n = 45,940 cells in Intermediate neuronal progenitors), represent IQR (25th, 50th, 75th percentile) with whiskers representing 1.5× IQR.