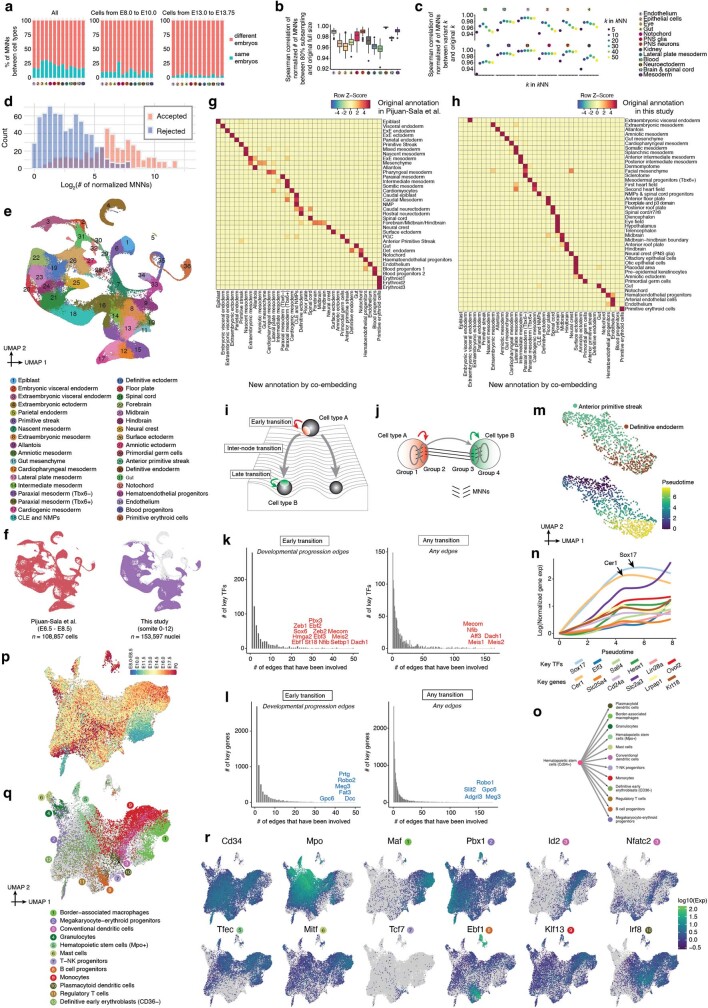
Extended Data Fig. 11. Identifying equivalent cell type nodes across datasets, and systematically nominating TFs and other genes for cell type specification.
a, The MNN approach used for graph construction is robust to subsampling and choice of the k parameter. The percentage of MNNs between different cell types, from the same embryo (blue) or from different embryos (red), is shown for each developmental system during organogenesis & fetal development, for all cells (left), cells from E8.0 to E10.0 (middle), or cells from E13.0 to E13.75 (right). b, The Spearman correlation coefficients of the normalized number of MNNs between cell types, comparing random subsampling of 80% of the cells to the full set of cells. The subsampling was repeated 100 times. The number of MNNs between cell types were normalized by the total number of possible MNNs between them. Boxplot (n = 1,200 correlation coefficients) represents IQR (25th, 50th, 75th percentile) with whiskers representing 1.5× IQR. Outliers are shown as the dots outside the whiskers. c, The Spearman correlation coefficients of the normalized number of MNNs between cell types, comparing various choices for k parameter (k = 5, 10, 20, 30, 40, 50) and the choice of k parameter (k = 15) when applying kNN to the developmental systems during organogenesis & fetal development. The number of MNNs between cell types were normalized by the total number of possible MNNs between them. Colors and numbers in panels a-c correspond to each developmental system annotations listed at the top right. d, 1,155 edges with the number of normalized MNNs > 1 were manually reviewed for biological plausibility. Histogram of edges that were accepted or rejected as a function of normalized MNN score. e, Integration of scRNA-seq profiles from gastrulation and early somitogenesis to identify equivalent cell type nodes across datasets generated by distinct technologies. 2D UMAP visualization of co-embedded cells, derived both from a gastrulation dataset based on cells from E6.5 to E8.5 generated on the 10x Genomics platform7 (n = 108,857 cells) and the earliest ~1% of this dataset (0-12 somite stage embryos) generated by sci-RNA-seq3 (n = 153,597 nuclei), after batch correction63. This is essentially an updated version of an analysis that we have done previously8. We performed clustering and cell type annotation on the integrated co-embedding, as shown. f, The same UMAP as in panel e is shown twice, with colors highlighting cells/nuclei from Pijuan-Sala’s dataset7 (left) or early somitogenesis8 (right). g, For cells from the original Pijuan-Sala’s dataset7, we quantify and display the overlap between the original annotations and the new annotations shown in panel e. For each row, the proportions of cells that are distributed across each column are transformed to z-score. h, For nuclei from the early somitogenesis embryos8, we quantify and display the overlap between the original annotations and the new annotations shown in panel e. These mappings were the basis for dataset equivalence edges between the “gastrulation” and 12 “organogenesis & fetal development” subsystems. For each row, the proportions of cells that are distributed across each column are transformed to z-score. CLE: Caudal lateral epiblast. NMPs: Neuromesodermal progenitors. i, A Waddington landscape cartoon illustrating how a cell type transition might be broken into three phases. j, Given a directional edge between two nodes, A → B, we identified the subset of cells within each node that were either MNNs of the other cell type (inter-node; groups 2 & 3) or MNNs of those cells (intra-node; groups 1 & 4). If A → B, this effectively models the transition as group 1 → 2 → 3 → 4. k, Histograms of the number of edges in which TFs are differentially expressed. The left histogram counts only genes when they are differentially expressed across the early phase of an developmental progression edge, while the right histogram counts genes when they are differentially expressed in any phase of all edges. l, Same as panel k, but for all genes rather than only TFs. m, Re-embedded 2D UMAP of 988 cells participating in groups 1-4 of the transition from anterior primitive streak → definitive endoderm. Cells are colored by either cell type annotations (top) or estimated pseudotime (bottom) using Monocle314. n, For cells in panel m, normalized gene expression of selected genes are plotted as a function of estimated pseudotime. Gene expression values were calculated from original UMI counts normalized to total UMIs per cell, followed by natural-log transformation. The line of gene expression was plotted by the geom_smooth function in ggplot2. We manually added an offset based on their expression at pseudotime = 0 to the y-axis for individual genes. o, A sub-graph of Fig. 5g, including hematopoietic stem cells (Cd34 + ) and 12 cell type nodes which appear derived from it. p, Re-embedded 2D UMAP of 37,750 cells from hematopoietic stem cells (Cd34 + ), colored by developmental stage (after downsampling to a uniform number of cells per stage). q, The same UMAP as in panel p, but with inferred progenitor cells (the cells participating in the MNNs that support the edges) colored by derivative cell type with the most frequent MNN pairs. r, The same UMAP as in panel p, colored by gene expression of selected top key TFs which were upregulated during the “early transition” for each derivative.