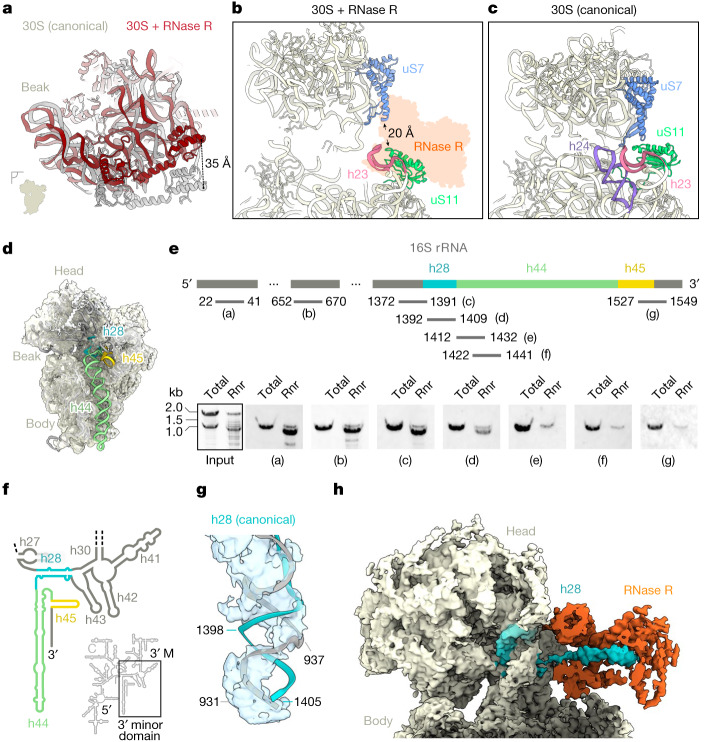
Fig. 2. RNase R is associated with a 30S degradation intermediate.
a, Comparison of the 30S head position between a canonical state (grey, Protein Data Bank (PDB) ID: 6HA8; ref. 33) and an RNase R-bound state I (dark red), when aligned on the 30S body. b,c, Relative positions of uS7 (blue) and uS11 (lime) with (b) and without (c) the presence of RNase R (PDB ID: 6HA8; ref. 33). Helix h24 (purple) is disordered and not modelled in the RNase R-bound state in c. d, Cryo-EM map of the RNase R-bound 30S fitted with the mature B. subtilis 30S model (PDB ID: 6HA8; ref. 33), showing a lack of density for h28 (turquoise), h44 (green) and h45 (yellow). The density has been filtered for visual clarity. e, Northern blot analysis of immunoprecipitated RNA with probes (designated by letter labels) as indicated in the schematic (n = 2). The schematic is not to scale. The input was stained with Serva stain G. Rnr, RNase R immunoprecipitated sample. f, Schematic of the secondary structure of B. subtilis 16S rRNA, highlighting the 3′ minor domain that contains h28 (turquoise), h44 (green) and h45 (yellow). 5′, 5′ domain; C, central domain, 3′ M, 3′ major domain. g, Cryo-EM map of the neck region of 30S (state I) with a fitted model of the canonical 16S rRNA (PDB ID: 6HA8; ref. 33). The density has been filtered for visual clarity. h, Cryo-EM map with an isolated density for RNase R (orange) and the h28 substrate (turquoise). The density has been filtered for visual clarity. For gel source data, see Supplementary Fig. 1.