FIGURE 1.
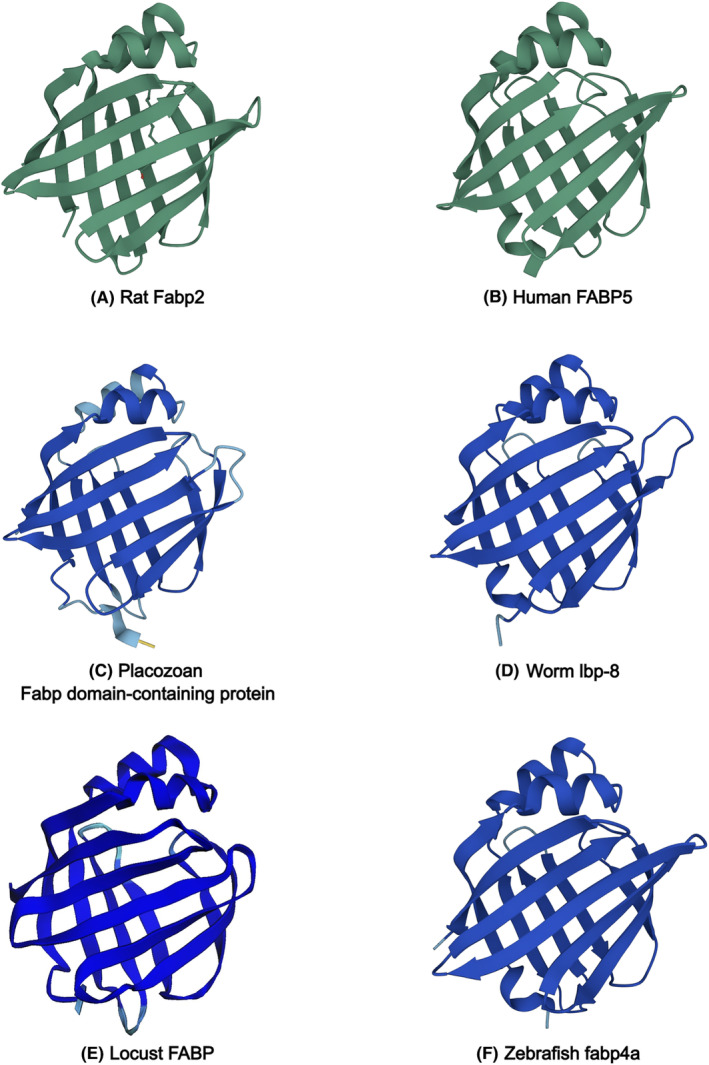
Structures of Fabps from different organisms determined experimentally (A, B) or predicted by AlphaFold 43 (https://alphafold.ebi.ac.uk/) (C–F). (A) Rat Fabp2 with bound ligand, determined by x‐ray crystallography (RCSB PDB 2IFB). (B) Human FABP5, determined by NMR spectroscopy (RCSB PDB 1JJJ). (C) Trichoplax adhaerens FABP domain‐containing protein (AlphaFold entry B3S4H2). (D) Caenorhabditis elegans lbp‐8 (AlphaFold entry O02324). (E) Schistocerca gregaria Fabp structure predicted from nucleotide sequence (NCBI accession XP_049839815.1) using AlphaFold2 via Colaboratory (https://colab.research.google.com/). (F) Danio rerio fabp4a (AlphaFold entry Q66I80).
