Figure 1: FKBP51 and FKBP52 Structure and Sequence Comparisons.
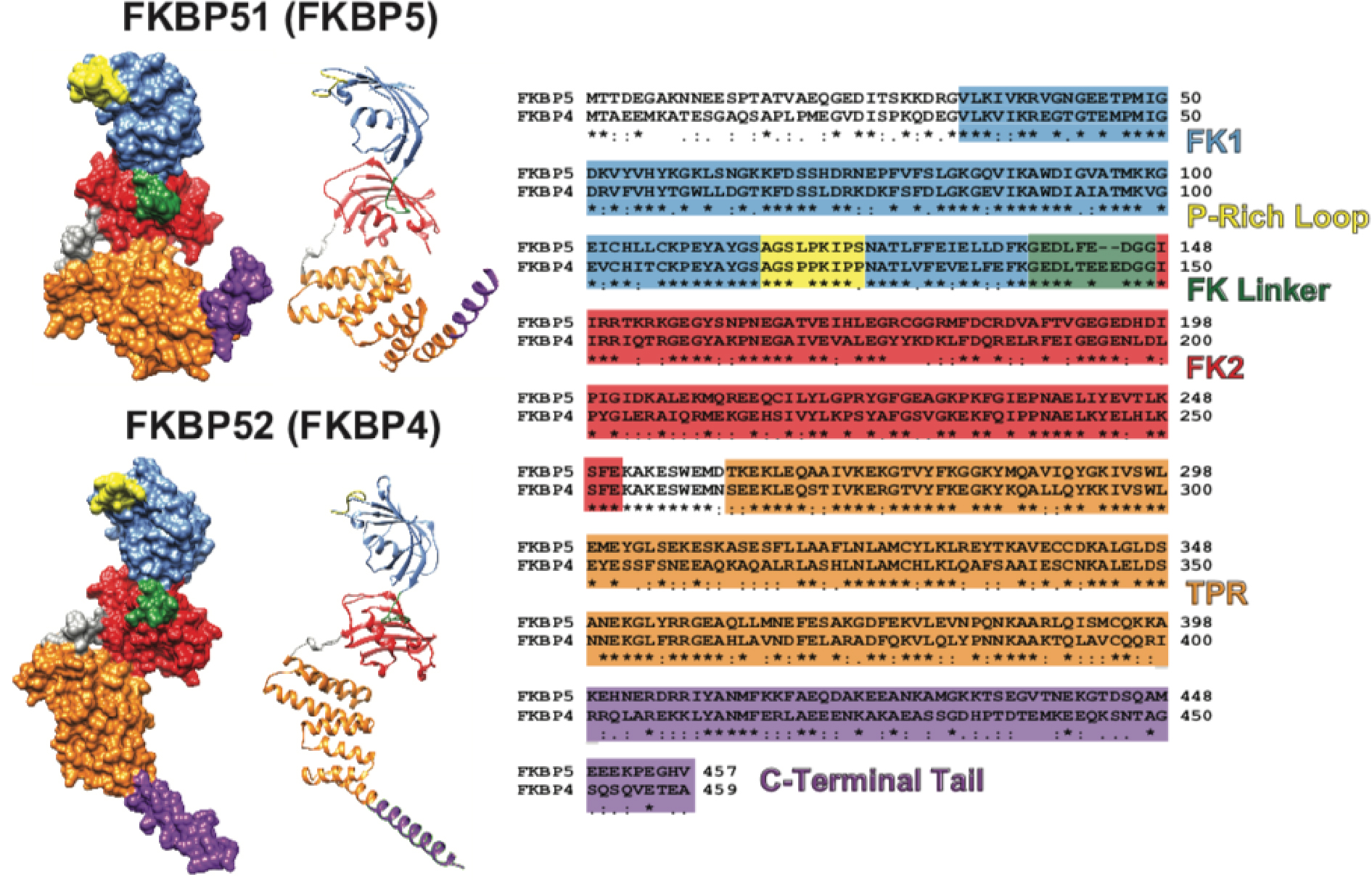
Both the ribbon and molecular surface models of the human FKBP51 crystal structure (FKBP5; PDB ID 1KT0) and overlaid images of two partial human FKBP52 crystal structures (FKBP4; PDB ID 1Q1C and 1P5Q) are shown on the left. The visible difference in the orientation of the TPR (orange) and C-Terminal Tail (purple) between FKBP51 and FKBP52 may be an artifact given that FKBP52 has been only partially crystalized. A multiple sequence alignment comparing the FKBP51 and FKBP52 amino acid sequence is shown on the right with known functional domains and regions colored according to their location on the crystal structures (* denotes identical amino acids, : denotes highly conserved amino acids, and . denotes partially conserved amino acids). Human FKBP51 and FKBP52 are approximately 60% identical and approximately 70% similar. The functional domains and regions highlighted include: the FK506 binding domain 1 (FK1) that contains a functional PPIase active site to which FK506 binds (blue); the proline-rich loop that overhangs the PPIase pocket and serves as a functionally relevant surface for the regulation of steroid hormone receptors (yellow); the linker region that links the two FK domains and contains a casein kinase 2 phosphorylation site (T143) in FKBP52 that may regulate Hsp90 binding (green); the FK2 domain that is structurally similar to FK1, but lacks a functional PPIase active site (red); the tetratricopeptide repeat (TPR) domain that mediates binding to the C-terminal EEVD motif on Hsp90 (orange); and the C-terminal Tail containing the Charge-Y motif that has also been shown to influence Hsp90 binding (purple). It is important to note that FKBP51 and FKBP52 were crystalized without the last 45 C-terminal amino acids and 32 C-terminal amino acids respectively. Thus, the structures shown on the left are truncated within the C-Terminal Tail. UCSF Chimera candidate version 1.12 (build 41600) was used to generate the ribbon and molecular surface images. The multiple sequence alignment was generated by CLUSTALW prior to illustration.
