Fig 4. Residual co-occurrence values and sequence patterns of Mastomys alleles found to be significantly associated with LASV infection.
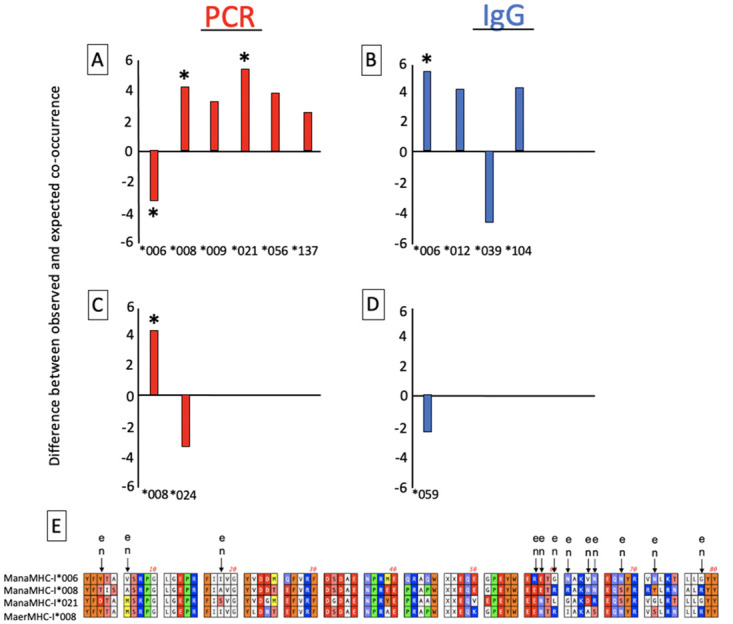
A) M. natalensis alleles significantly associated with LASV PCR. B) M. natalensis alleles significantly associated with LASV IgG. C) M. erythroleucus alleles significantly associated with LASV PCR. D) M. erythroleucus alleles significantly associated with LASV IgG. Asterisks denote significant LASV-allele relationships from both the co-occurrence analyses and General Linear Mixed Effect Models (GLMMs). Mana*009, *056 and *137 were correlated with *008; Mana*012, *039 and *104 were correlated with *006; and Maer*024 was correlated with Maer*008; In this case, GLMMs were only computed for the more frequent alleles (see Methods). E) Amino acid sequences of alleles significantly associated with LASV occurrence in both the co-occurrence analyses and GLMMs, showing positively selected sites common to both M. natalensis (n) and M. erythroleucus (e).
