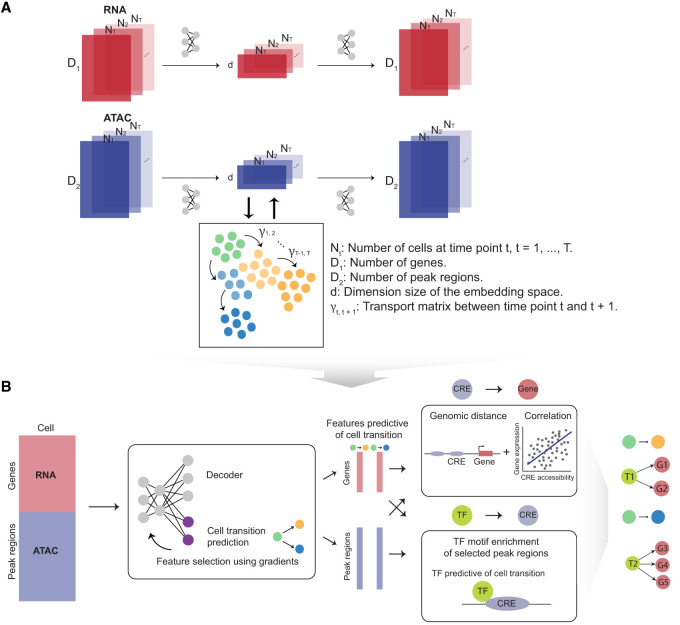
Figure 1.
Overview of scTIE, a unified framework for the integration of temporal data and the inference of context-specific GRNs that predict cell fates. The input of scTIE consists of the gene expression matrix of scRNA-seq and peak matrix of scATAC-seq from single-cell multiome data over a time course. scTIE consists of two main steps. (A) In the first step, each cell, represented by a pair of gene and peak vectors, is projected into a common embedding space by separate encoders and decoders. The two modalities and time points are aligned by appropriate loss functions, whereas the transition probability matrix between cells from consecutive time points is iteratively estimated. (B) In the second step, users have the ability to select specific subgroups of cells whose transitions are of interest, finetune the previously trained neural network, identify features that are predictive of transition probabilities, and construct the corresponding GRNs.