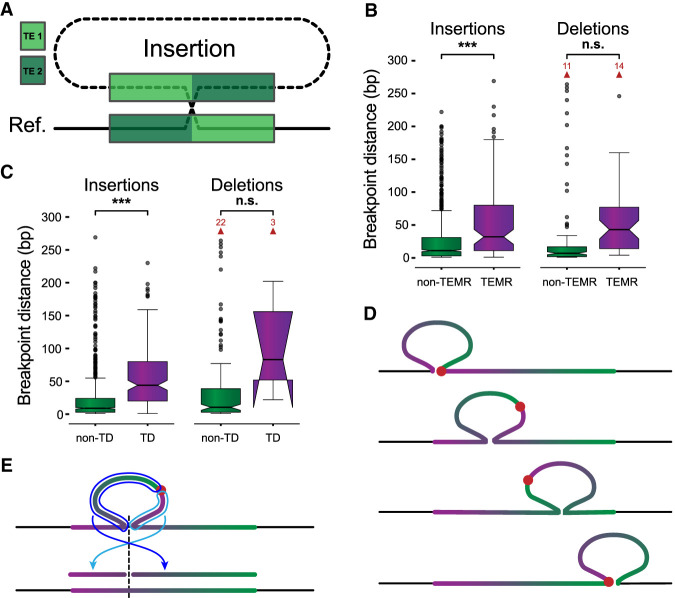
Figure 2.
Breakpoints shift through homology and alter SV representation. (A) A nonreference sequence with chimeric TE copies (TE 1, light green; TE 2, dark green) at the breakpoints. (B) The maximum distance between differential breakpoints across haplotypes is greater in TEMR variants with a significant difference for insertions. (C) Maximum breakpoint offsets for non-TD (green) and TD (violet) SVs. Horns extending downward on TD deletions indicate that the 95% confidence interval for the median extends below the bottom quartile. (D) The sequence of a tandem duplication shifts with different alignment positions along the reference copy. The junction of the duplication copies (red dot) is located at the insertion breakpoint if the insertion is placed at the left or right end of the reference copy (top and bottom examples); otherwise, the junction is embedded within the insertion creating a chimera of duplication copies inside the SV insertion (middle examples). (E) Mapping a chimeric TD to the reference occurs in two pieces separated at the TD breakpoint (light and dark blue arrows), where each piece maps to one side of the reference insertion site (dashed line). Alignment programs often miss one or both fragments. (B,C) P-values are generated from t-tests of the mean. Notches indicate a 95% confidence interval around the median. Red arrows and numbers indicate the number of outlier points above the horizontal axis maximum. (n.s.) Not significant, (*) 1 × 10−3 < P ≤ 1 × 10−2, (**) 1 × 10−4 < P ≤ 1 × 10−3, (***) P ≤ 1 × 10−4.