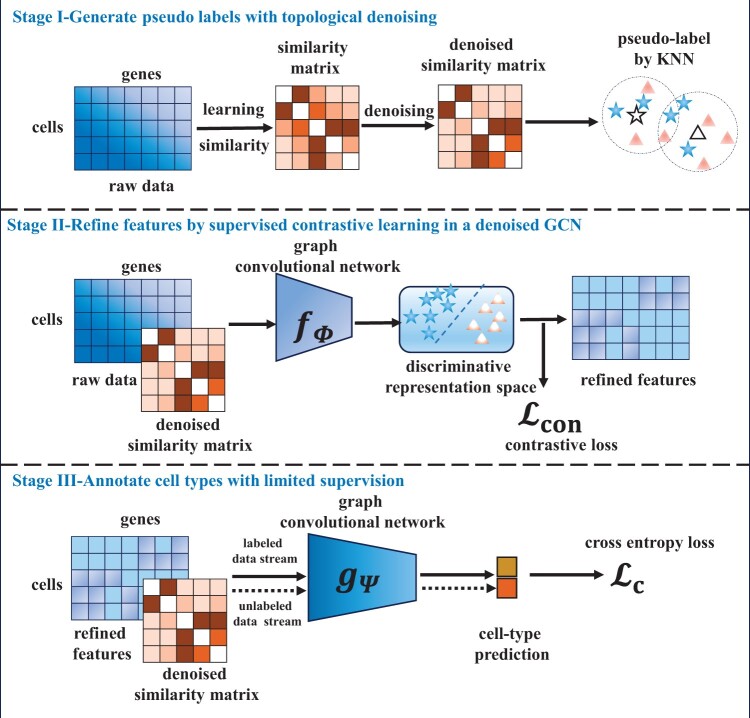
Figure 1.
Framework of scSemiGCN. It consists of three stages: (i) generating pseudo labels for unannotated cells with denoised similarities in k-nearest neighbors (KNN); (ii) projecting raw features onto a discriminative representation space by supervised contrastive learning; (iii) training a cell-type annotation model with labeled cells in a two-layer graph convolutional network (GCN) using refined features and the denoised network structure as inputs.