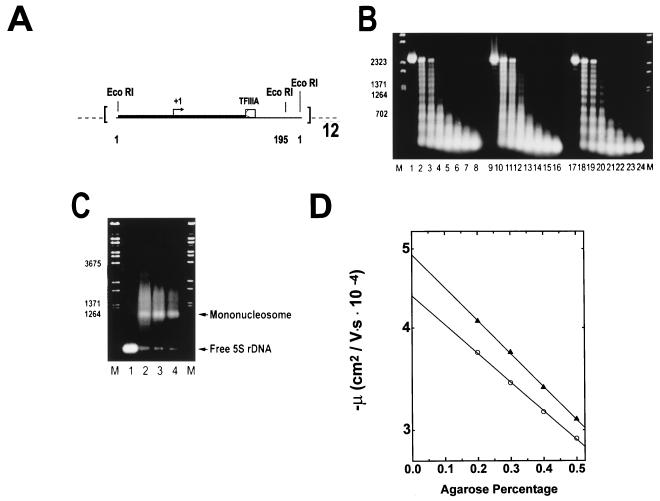
FIG. 2.
Reconstitution of underacetylated, moderately acetylated, and highly acetylated 208-12 nucleosomal arrays. (A) Schematic illustration of the 208-12 DNA template used for reconstitution. The 208-12 DNA consists of 12 tandem repeats of a portion of the Lytechinus 5S rRNA gene (51). Each 5S rDNA repeat contains both a preferred nucleosome positioning site (solid box) and a TFIIIA binding site (open box). Initiation of transcription by RNA polymerase III occurs ∼90 bp upstream of the major TFIIIA binding site (+1, arrow). The termination sequence of the 5S rRNA gene was deleted during template construction (51), allowing for production of long read-through transcripts. EcoRI digestion sites are located at sequences 1 and 195 of each 5S rDNA repeat. (B) Micrococcal nuclease digestion. Underacetylated (lanes 1 to 8), highly acetylated (lanes 9 to 16), and moderately acetylated (lanes 17 to 24) reconstitutes were digested with 0.05 U of micrococcal nuclease per μg of DNA in the presence of 1 mM CaCl2. Digestion was for 0, 0.5, 1.0, 2.5, 5, 10, 20, and 30 min (shown from left to right for each reconstitute type) at room temperature. The reactions were quenched by addition of a 1/5 volume of 5% SDS–25% glycerol–10 mM EDTA–0.3% bromophenol blue. Samples were deproteinated by incubation at 37°C for 30 min and subsequently electrophoresed for 5 h at 2 V/cm in a 1% agarose gel buffered with 40 mM Tris-acetate–1 mM EDTA (pH 8.0). Bands were visualized under UV illumination after incubation of the gel in ethidium bromide. Lambda DNA digested with BstEII (lanes M) was used for the size markers. (C) EcoRI digestion. One microgram (each) of naked 208-12 DNA (lane 1) and underacetylated, highly acetylated, and moderately acetylated nucleosomal arrays (lanes 2 to 4, respectively) was digested with 10 U of EcoRI for 60 min at room temperature in digestion buffer H (Promega). Reactions were quenched by addition of a 1/5 volume of 25% glycerol–10 mM EDTA (pH 8.0). Digestion products were electrophoresed at 4 V/cm for 3 h in a 1% agarose gel buffered with 40 mM Tris-acetate–1 mM EDTA (pH 8.0). Lambda DNA digested with BstEII (lanes M) was used for the size markers. (D) Determination of μ0. Shown are plots of the mobilities in 0.2 to 0.5% agarose of underacetylated (○) and highly acetylated (▴) nucleosomal arrays in E buffer. The μ0 was determined from the extrapolated gel-free mobilities as described in Materials and Methods.