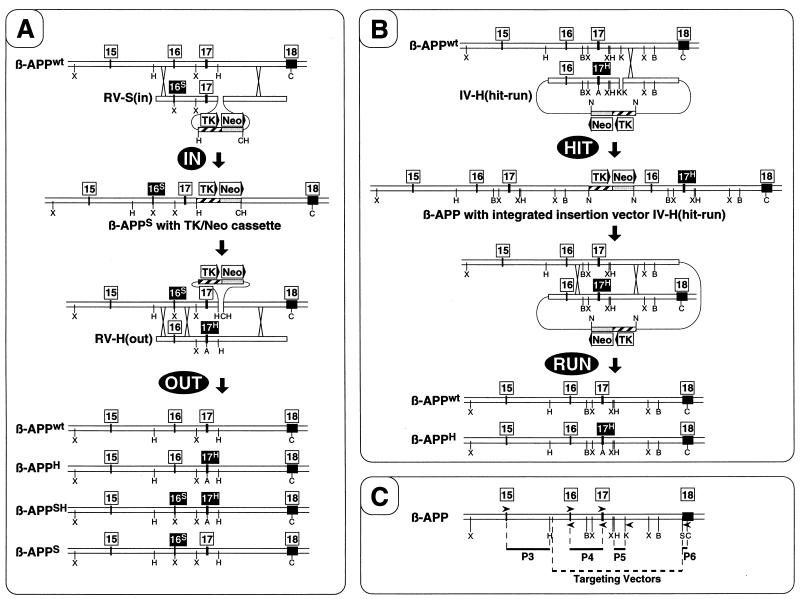
FIG. 1.
Approaches for introduction of subtle mutations into ES cells by homologous recombination. (A) The targeting-in-out procedure. Boxes with numbers indicate the positions of exons. Those exons carrying the SFAD (16S) or HCHWA-D (17H) mutation are highlighted, and mutated β-APP alleles are labeled by the same superscript letters. Boxes with text and arrowheads indicate the positions and orientations of selection genes. The locations of crossovers during homologous recombination are indicated by crosses. After the first homologous recombination (targeting IN), ES cells having replaced the targeted locus by the replacement vector RV-S(in) are enriched by selection for neomycin resistance (Neo) with G418. Isolated targeted-in ES cells are subsequently targeted OUT again by homologous recombination with RV-H(out) and selection with FIAU against the viral TK. Four possible end arrangements are shown, three of which contain mutations but lack the selection cassette. (B) The hit-and-run procedure. Selection of hit clones with a partial genomic duplication due to targeted integration of the vector IV-H(hit-run) by homologous recombination is performed as in the targeting-in step. After the run, ES cells that had undergone spontaneous intrachromosomal recombination resulting in loss of the duplication and selection cassette are counterselected with FIAU against TK. This process ends up either in reversion to wild-type (wt) or in expected targeted mutated ES cells without a selection cassette. (C) Primers and probes. Probes 3 to 6 (P3 to P6) for genomic Southern blot analysis are indicated by dark solid lines, and the genomic fragment used for construction of targeting vectors is shown as a dotted line. The binding sites and 5′-to-3′ orientation of primers used for PCR are shown by arrowheads. The positions of recognition sites for restriction endonucleases are indicated by single letters: A, AatII; B, BamHI; C, ClaI; H, HindIII; K, KpnI; N, NotI; S, SalI; X, XbaI.