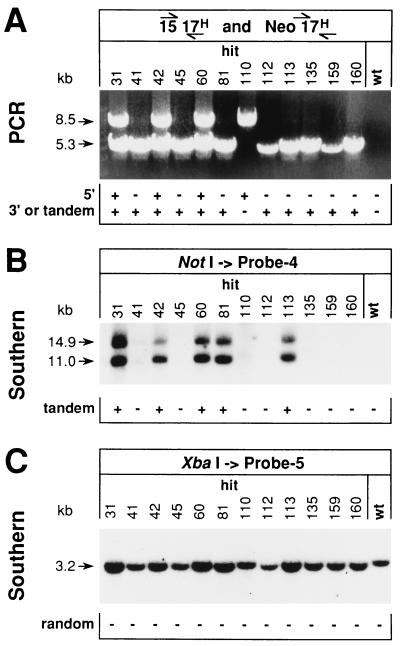
FIG. 4.
Characterization of potential hit ES cells. (A) Analysis of the targeted locus of hit clones. The position of the HCHWA-D mutation on the partial genomic duplication of hit clones was investigated by two mutation-specific PCRs by using primer pairs 15F–17R-H for detection of the 5′ location and NeoF–17R-H for the 3′ location. A combination of 15-μl aliquots from both reaction mixtures was analyzed directly by electrophoresis on 0.6% agarose gels. The band at 8.5 kb indicates the site-specific 5′ position of the HCHWA-D mutation, and the 5.3-kb band indicates the mutation when located 3′ of the selection cassette. (B) Detection of possible repetitive integrations of the targeting vector by Southern blot analysis. Genomic DNA isolated from positive ES cell clones was digested with NotI, separated by electrophoresis on 0.6% agarose gels, and hybridized with intron 16-specific probe 4 after transfer to a nitrocellulose membrane. The appearance of a band at 11 kb indicated the presence of head-to-tail tandem repeats of IV-H(hit-run) in some clones. Because of incomplete NotI restriction, a 14.9-kb band was detected together with the expected 11-kb band, showing elongated fragments, i.e., products with an attached selection cassette. (C) Exclusion of additional random integrations of targeting vectors. XbaI-restricted DNA was run on 0.9% agarose gels and tested with intron 17-specific probe 5. Except for the 3.2-kb band for the wild type and for alleles with site-specific integrations of IV-H(hit-run), no additional bands indicative of random integrations were detected. However, because of the higher copy number, stronger 3.2-kb signals were obtained from those clones with specifically integrated vector repeats. wt, wild type. Numbers at tops of lanes are clone designations.