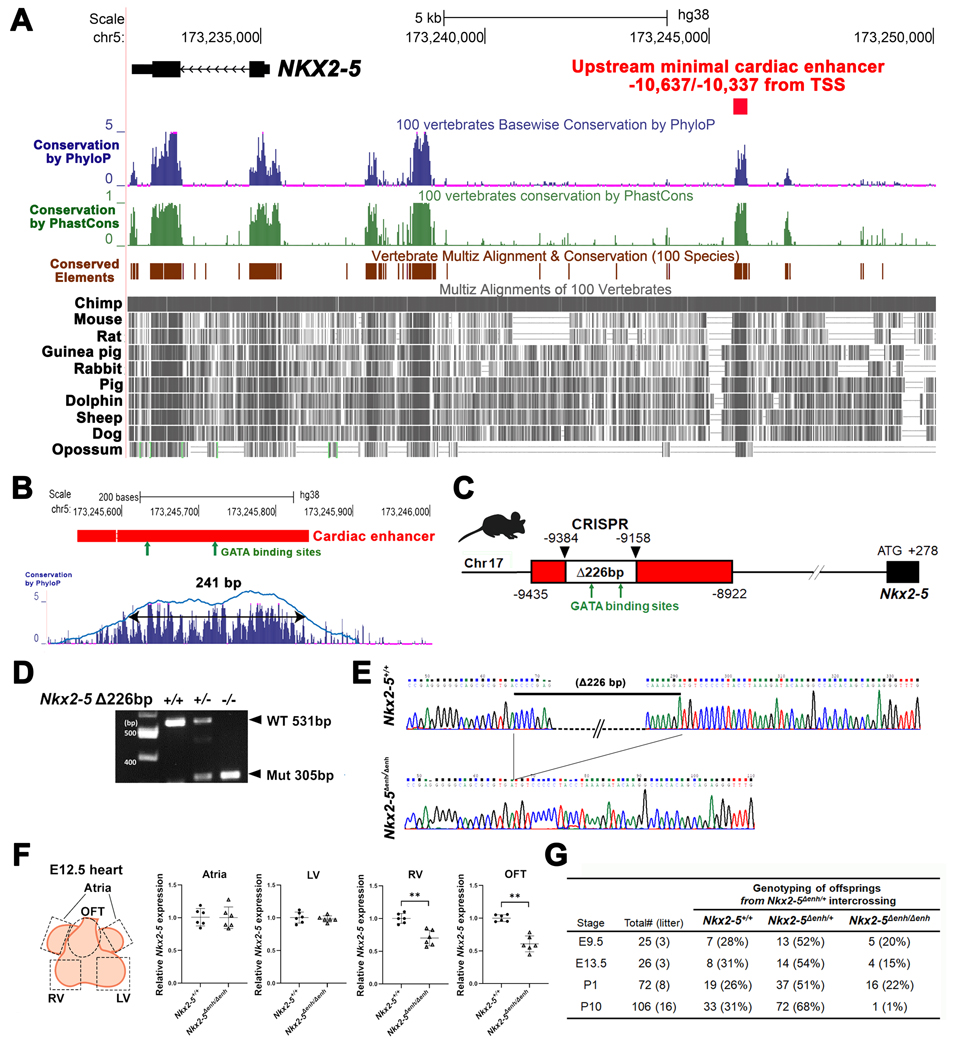
Figure 1. Generation of Nkx2–5∆enh/∆enh mice engineered with a 226-bp deletion in a highly conserved enhancer region.
(A) Conservation state of the upstream NKX2–5 cardiac enhancer by the UCSC Genome Browser. NKX2–5 coding sequence (labeled in black) and an upstream minimal cardiac enhancer region (labeled in red) on the human hg38 assembly. Conservation score using phyloP (blue bars) and phastCons (green bars) reveals that sequences corresponding to the cardiac enhancer are highly conserved among vertebrates. Conserved elements identified by phastCons are also displayed in brown. Representative vertebrate genomes of selected species are displayed in gray. TSS, transcription start site. (B) A 301-bp region (labeled in red) on human hg38 assembly with 90% sequence similarity to the previously reported cardiac enhancer region on the mouse mm10 assembly with conserved GATA-binding sites (green arrows)(upper panel). A gap with double horizontal lines in the cardiac enhancer shows dissimilarity in sequence between human and mouse genomes. Further restriction of the conservation peaks above 50% threshold yielded a 241-bp region containing the two GATA-binding sites (lower panel) (C) Schematic of CRISPR-CAS9 deletion of a 226-bp fragment containing two conserved GATA-binding sites ≈9.4 kb upstream of the Nkx2–5 transcription start site in a mouse model with mm10 assembly. (D) PCR of genomic DNA from wildtype, heterozygous, and homozygous cardiac enhancer deletion (Nkx2–5∆enh) mice. (E) Sequencing of genomic DNA from wildtype Nkx2–5+/+ and homozygous Nkx2–5∆enh/∆enh mice showing deletion of the 226-bp fragment. (F) RT-qPCR from regional heart tissues at embryonic day E12.5 demonstrating reduced Nkx2–5 expression exclusively in the right ventricle (RV) and outflow tract (OFT). LV: left ventricle. Groups (n = 6 each) were compared by Student’s t-test. **p < 0.01. Data are presented as mean ± SD. (G) Genotype of viable offspring at each developmental stage from Nkx2–5∆enh/+ mating pairs. Number of viable Nkx2–5∆enh/∆enh at postnatal day (P) 10 is significantly lower than the expected Mendelian distribution.