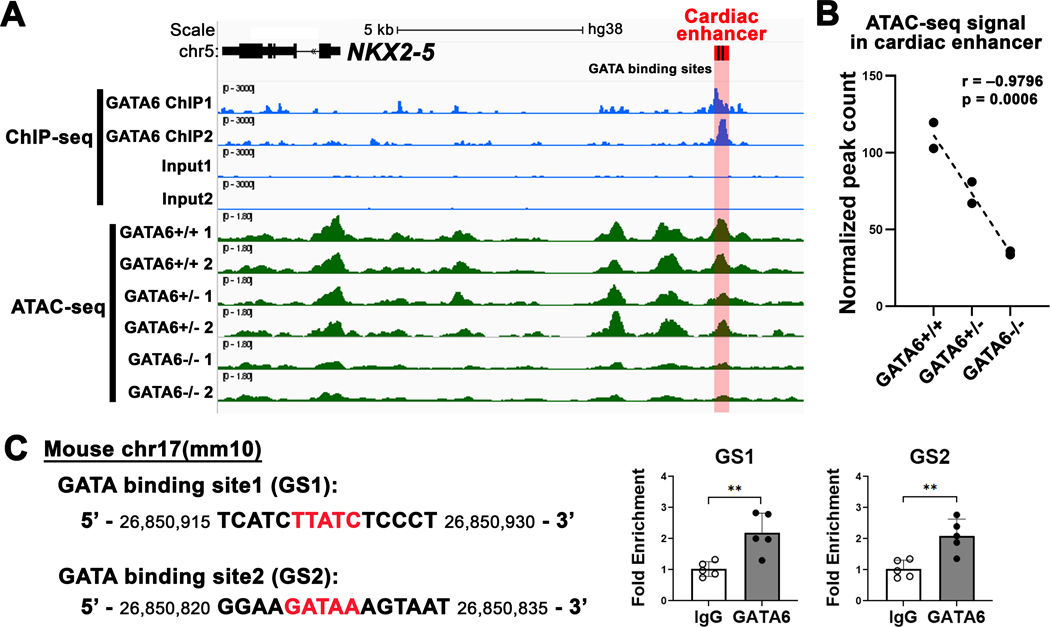
Figure 7. Endogenous GATA6 interacts with the NKX2–5 cardiac enhancer and determines enhancer accessibility in human cardiomyocytes.
(A) Visualization of GATA6 chromatin immunoprecipitation sequencing (ChIP-seq) and assay of transposase accessible chromatin sequencing (ATAC-seq) datasets in human-induced pluripotent stem cell derived cardiomyocytes (hiPSC-CMs). ChIP-seq for GATA6 and input control in day 4 hiPSC-CMs in blue. ATAC-seq in day 4 GATA6 isogenic wildtype (GATA6+/+) and mutants (GATA6+/− and GATA6−/−) in green. The conserved cardiac enhancer region shown in red containing two GATA-binding sites in black. A GATA6 ChIP-seq peak is identified in the cardiac enhancer region (adjusted p value < 0.05 by peak-calling analysis). Differential ATAC-seq peak analysis of the cardiac enhancer region based on GATA6 genotype. (B) Pearson correlation between normalized peak count in the cardiac enhancer region and experimental groups for ATAC-seq. GATA6 mutant allele correlates with decreased ATAC-seq peak counts. (C) ChIP-quantitative PCR (ChIP-qPCR) validation of GATA6 binding in embryonic day E12.5 mouse hearts. Conserved GATA-binding sites (red) in the cardiac enhancer region on the mouse mm10 assembly (left panel). Independent ChIP-qPCR for two GATA-binding sites, GS1 and GS2, in E12.5 mouse hearts showing GATA6 occupancy for both sites. **P < 0.01 by Student’s t-test. Data are presented as mean ± SD. n = 5 per group.