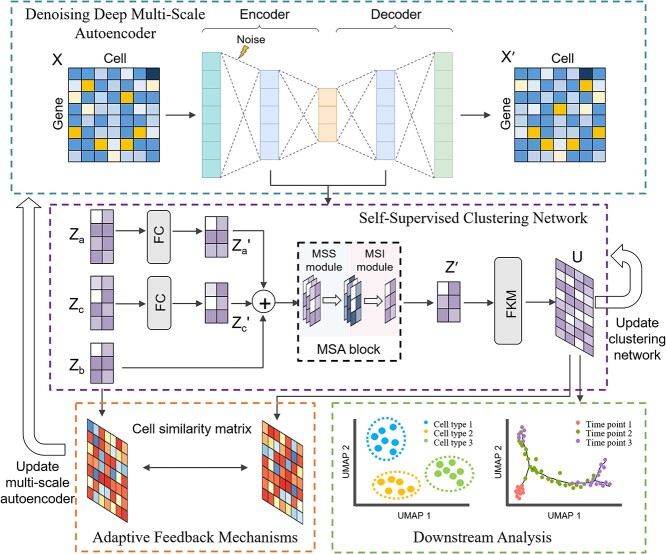
Figure 1.
Overall architecture diagram of scAMAC. scAMAC consists of two parts: denoising deep multi-scale autoencoder and self-supervised clustering network. Firstly, the preprocessed gene expression matrix 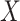 is fed into the denoising deep multi-scale autoencoder, obtaining the latent feature representation
is fed into the denoising deep multi-scale autoencoder, obtaining the latent feature representation 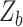 of the hidden layer, as well as the reconstructed data
of the hidden layer, as well as the reconstructed data 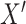 . Then, the outputs of the encoding layer and decoding layer are fed into two fully connected layers to obtain
. Then, the outputs of the encoding layer and decoding layer are fed into two fully connected layers to obtain 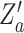 and
and 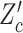 , where the dimensions of
, where the dimensions of 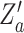 and
and 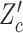 are the same as
are the same as 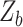 . Finally,
. Finally, 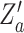 ,
, 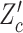 , and
, and 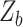 are concatenated and fed into the self-supervised clustering network. The self-supervised clustering network uses the MSA mechanism to capture the relationship between cells and the contribution of each layer of the autoencoder to obtain
are concatenated and fed into the self-supervised clustering network. The self-supervised clustering network uses the MSA mechanism to capture the relationship between cells and the contribution of each layer of the autoencoder to obtain 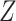 . The membership matrix
. The membership matrix 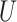 of the FKM algorithm is calculated based on
of the FKM algorithm is calculated based on 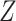 and optimized for the self-supervised clustering network. Meanwhile,
and optimized for the self-supervised clustering network. Meanwhile, 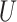 is used to construct a cell similarity matrix to supervise the parameter update of the autoencoder.
is used to construct a cell similarity matrix to supervise the parameter update of the autoencoder.