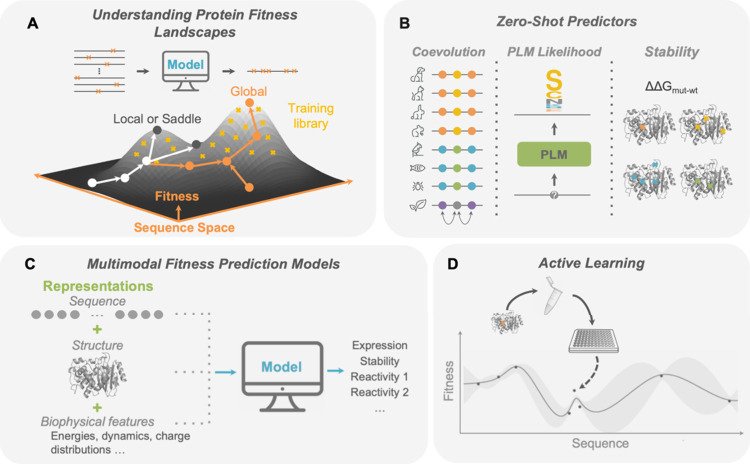
Figure 3.
Opportunities for machine learning models to help navigate protein fitness landscapes. (A) ML models can allow for bigger jumps in sequence space by proposing combinations of mutations that would not be achieved by traditional DE. The role of nonadditivity between mutation effects, or epistasis, should be explored further to understand when ML offers an advantage. (B) The role of ZS scores to predict protein fitness without any labeled assay data needs to be better understood for different protein families and functions. Finally, ML-assisted protein fitness optimization could benefit from (C) multimodal representations that capture physically relevant descriptors of proteins to predict multiple relevant properties and (D) active learning with deep learning models tailored toward proteins and uncertainty quantification.