Abstract
Parkinson’s disease is an age-related neurodegenerative disorder with a higher incidence in males than females. The causes for this sex difference are unknown. Genome-wide association studies (GWAS) have identified 90 Parkinson’s disease risk loci, but the genetic studies have not found sex-specific differences in allele frequency on autosomal chromosomes or sex chromosomes. Genetic variants, however, could exert sex-specific effects on gene function and regulation of gene expression.
To identify genetic loci that might have sex-specific effects, we studied pleiotropy between Parkinson’s disease and sex-specific traits. Summary statistics from GWASs were acquired from large-scale consortia for Parkinson’s disease (n cases = 13 708; n controls = 95 282), age at menarche (n = 368 888 females) and age at menopause (n = 69 360 females). We applied the conditional/conjunctional false discovery rate (FDR) method to identify shared loci between Parkinson’s disease and these sex-specific traits. Next, we investigated sex-specific gene expression differences in the superior frontal cortex of both neuropathologically healthy individuals and Parkinson’s disease patients (n cases = 61; n controls = 23). To provide biological insights to the genetic pleiotropy, we performed sex-specific expression quantitative trait locus (eQTL) analysis and sex-specific age-related differential expression analysis for genes mapped to Parkinson’s disease risk loci.
Through conditional/conjunctional FDR analysis we found 11 loci shared between Parkinson’s disease and the sex-specific traits age at menarche and age at menopause. Gene-set and pathway analysis of the genes mapped to these loci highlighted the importance of the immune response in determining an increased disease incidence in the male population. Moreover, we highlighted a total of nine genes whose expression or age-related expression in the human brain is influenced by genetic variants in a sex-specific manner. With these analyses we demonstrated that the lack of clear sex-specific differences in allele frequencies for Parkinson’s disease loci does not exclude a genetic contribution to differences in disease incidence. Moreover, further studies are needed to elucidate the role that the candidate genes identified here could have in determining a higher incidence of Parkinson’s disease in the male population.
Keywords: conjunctional FDR, brain sex-specific gene expression, sex-specific eQTLs, sex-specific age-related gene expression
Parkinson’s disease has a higher incidence in males than females, but the causes of this sex difference are unknown. Nordengen et al. identify eleven genomic loci shared between Parkinson´s disease and sex-specific traits, and highlight nine genes whose expression is influenced by genetic variants in a sex-specific manner.
Introduction
Parkinson’s disease is an age-related, progressive neurodegenerative disorder that affects 2–3% of the world population over 65 years of age.1 Epidemiological studies have consistently found that the incidence of Parkinson’s disease is around 1.5 times higher in males than females.2 The causes for the observed increased risk in males compared to females is still poorly understood. Various mechanisms have been proposed to explain this sex difference in Parkinson’s disease risk, including different degrees of exposure to environmental factors, such as pesticides or smoke, and the effect of sex hormone levels and other genetic factors influenced by biological sex.3-5
Three recent studies examined the genetic components to this unexplained sex difference.6-8 An X-chromosome-wide association study identified two novel genetic risk loci for Parkinson’s disease and a significant expression quantitative trait locus (eQTL) for one of them but found no significant differences between males and females.6 Further, Blauwendraat and colleagues7 performed a sex-stratified genome-wide association study (GWAS) in Parkinson’s disease and reported no sex differences for autosomal genetic variation. Additionally, an analysis of Y-chromosome haplogroups failed to find strong association between Y-chromosome variants and Parkinson’s disease.8 These studies found no support for the hypothesis that sex differences in allele frequency of genetic risk variants explain the difference in Parkinson’s disease incidence.
This, however, does not exclude a genetic contribution to differences in Parkinson’s disease incidence. Genetic variants present in both sexes may not have identical biological effects, for example due to the interaction between the genetic variants and sex hormones signalling pathways. Exposure to oestrogens, the female sex hormones, has been highlighted as a potential cause of the lower Parkinson’s disease incidence in females. In experimental studies, oestrogens have been shown to upregulate neurotrophic factors,9 increase dopamine synthesis,9 decrease inflammation10 and prevent α-synuclein aggregation and Lewy body formation.11 In accordance with this, an association between factors reducing oestrogen stimulation during life and Parkinson’s disease has been found,12-14 including short fertile life length,12 early menopause through bilateral oophorectomy13 and other fertile life characteristics. Equivalent with this, the genes affecting length of oestrogen exposure through life (early menarche and late menopause) might also affect sex-specific disease risk.
Furthermore, sex-specific gene regulatory structures exist in the human brain.15,16 Supporting this notion, a study conducted in several human brain regions showed that GWAS hits can be associated with variation in gene expression in one sex, but not the other. This demonstrates the existence of sex-specific eQTLs.17 Moreover, age is also known to influence gene expression in a sex-specific manner.18 Similar studies investigating sex-specific eQTLs or sex-specific age-related expression have never been conducted using Parkinson’s disease GWAS.
We approached the question of possible genetic components to sex differences in Parkinson’s disease by examining pleiotropy between Parkinson’s disease and sex-specific traits. Pleiotropy occurs when one genetic variant influences two or more seemingly unrelated phenotypic traits. We took advantage of summary statistics from GWAS of Parkinson’s disease19 and sex-specific traits to study genetic pleiotropy between Parkinson’s disease and the two traits age at menarche20 and age at menopause.21 We choose these sex-specific traits because they are both influenced by oestrogen levels, and oestrogens have been implicated in Parkinson’s disease pathogenesis.22-32 Furthermore, to investigate the potential of sex-specific translational changes among the identified Parkinson’s disease risk loci, we analysed sex-specific gene expression in the frontal cortex of neuropathologically healthy donors and Parkinson’s disease patients. Finally, we used our gene expression data to prioritize genes in the identified pleiotropy loci and in previously identified Parkinson’s disease loci.
Materials and methods
Genome wide association study samples
We acquired GWAS summary statistics for Parkinson’s disease excluding 23andMe, Inc., comprising 13 708 cases and proxy-cases and 95 282 controls.19 The age at menarche sample including 23andMe, Inc. comprised 368 888 females20 and the age at menopause sample comprised 69 360 females.33 Studies were published between July 2014 and December 2019 and selected to maximize available sample size. Full genotyping procedures are detailed in the original publications.
Genetic pleiotropy analyses
To visually assess the presence of enrichment, we generated conditional quantile-quantile (Q-Q) plots34,35 conditioning age at menarche and age at menopause on Parkinson’s disease (Fig. 1A). To determine any loci likely to be shared by two phenotypes, we computed conjunctional false discovery rate (FDR) statistics.35 The conjunctional FDR is an extension of the conditional FDR (see the Supplementary material for detailed description) and is defined as the maximum of the two conditional FDR statistics for a specific single nucleotide polymorphism (SNP) and estimates the posterior probability that a SNP is null for either trait or both, given that the P-values for both phenotypes are as small as or smaller than the P-values for each trait individually. For more details, see the original34,36 and subsequent publications.37-39 The threshold for significance was set at FDR < 0.05. Manhattan plots (Fig. 2) were constructed based on the conjunctional FDR to show the genomic localization of the shared genetic loci. We excluded all SNPs from the major histocompatibility complex (MHC) region (chromosome 6: 25 119 106–33 854 733), the chromosome 8: 7 242 715–12 483 982 region and the MAPT region (chromosome 17: 40 000 000–47 000 000), as these regions are known to exhibit high linkage disequilibrium (LD) and the inclusion of SNPs in high LD can lead to inflation of pleiotropy analysis.
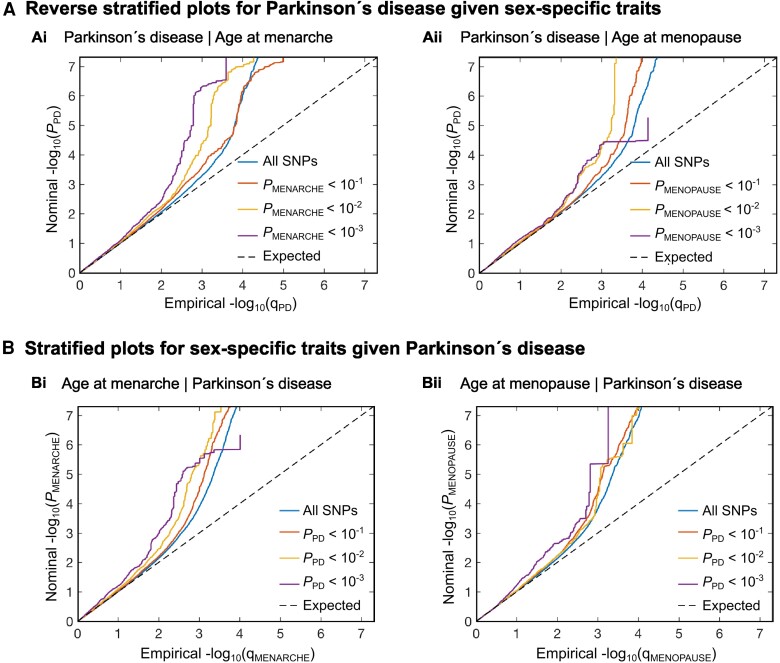
Figure 1.
Cross-trait enrichment between Parkinson’s disease and sex-specific traits. (A) The reverse stratified quantile-quantile (Q-Q) plots show successive increments for single nucleotide polymorphism (SNP) enrichment for [A(i)] age at menarche and [A(ii)] age at menopause conditional on SNP associations with Parkinson’s disease. Conditional Q-Q plots of nominal versus empirical −log10 P-values (corrected for inflation) in Parkinson’s disease below the genome-wide association study (GWAS) threshold of P < 5 × 10−8 as a function of significance of association with the sex-specific traits, at the level of P < 0.1, P < 0.01 and P < 0.001. The blue lines indicate all SNPs. The dashed lines indicate the null hypothesis. (B) The stratified Q-Q plots show successive increments for SNP enrichment for Parkinson’s disease conditional on SNP associations with [B(i)] age at menarche and [B(ii)] age at menopause. Conditional Q-Q plots of nominal versus empirical −log10 P-values (corrected for inflation) in Parkinson’s disease below the GWAS threshold of P < 5 × 10−8 as a function of significance of association with sex-specific traits at the level of P < 0.1, P < 0.01 and P < 0.001.
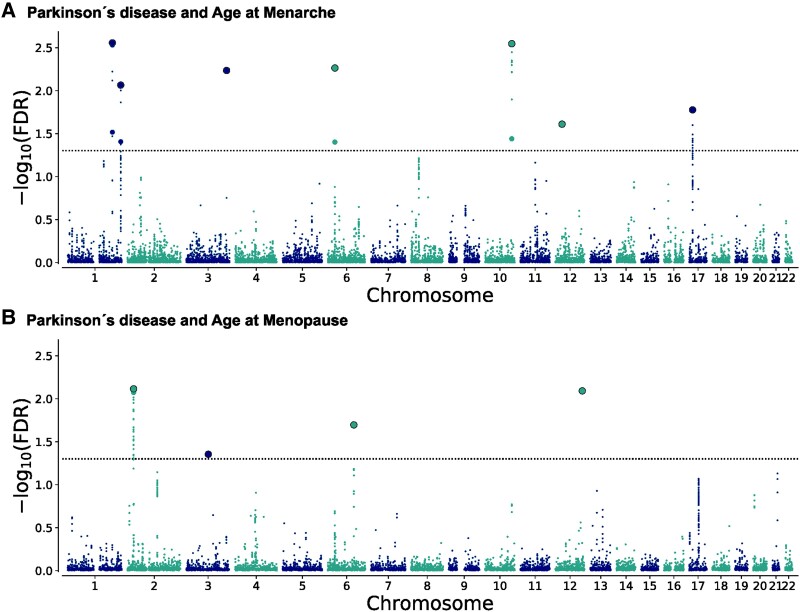
Figure 2.
Chromosomal distribution of genetic loci shared between Parkinson’s disease and sex-specific traits. The Manhattan plots illustrate the chromosomal distribution of shared genetic loci between Parkinson’s disease and (A) age at menarche and (B) age at menopause. The −log10 transformed conjunctional false discovery rate (conjFDR) values for each single nucleotide polymorphism (SNP) are shown on the y-axis against chromosomal position on the x-axis. The dashed lines represent the conjFDR threshold for significant association (FDR < 0.05). Black outlined circles represent independent lead SNPs.
Genetic loci definition and functional annotation
We functionally annotated all candidate SNPs of the shared loci between age at menarche and age at menopause with Parkinson’s disease using FUMA.40 SNPs with FDR less than 0.01 and LD r2 < 0.6 with each other were considered as independent significant SNPs, and a fraction of the independent significant SNPs in approximate LD with each other at r2 < 0.1 were considered lead SNPs. We outlined the distinct genomic loci and their borders based on FUMA’s default parameters.40
FUMA was also deployed to annotate the significant associated SNPs with functional categories, combined annotation dependent depletion scores (CADD),41 RegulomeDB scores (RBD)42 and chromatin states.43,44 A CADD score above 12.37 shows an association of deleterious protein with outcomes.41 The RBD score indicates the regulatory functionality of SNPs based on eQTL and chromatin marks.42 The chromatin state indicates the accessibility of genomic regions using 15 categories, as predicted by ChromHMM based on five chromatin marks for 127 epigenomes.43,44 To place them in potential biological context, we matched the candidate loci to eQTL databases from GTEx (http://gtexportal.org) from brain, blood and ovaries and Braineac (http://www.braineac.org).45
On each set of mapped genes, we performed gene-set enrichment analysis within the Gene Ontology classification system.46,47 We tested for overrepresentation of mapped genes within pathways derived from 12 public resources, collated by ConsensusPathDB and corrected for multiple testing using the q-value.48 We also constructed spatiotemporal heat maps of gene expression levels across 11 brain regions at 11 developmental time points with the R package ‘cerebroViz’ using BrainSpan RNA sequencing data.49-51 Expression across brain tissues was clustered using unsupervised hierarchical cluster analysis.
Post-mortem cohort
The 84 individuals included for the brain gene expression analyses were selected based on neuropathologically post-mortem characterization performed by an experienced neuropathologist (A.J.M.R.) or neuroanatomist (W.D.J.v.d.B), including assessment of Lewy body-related α-synuclein pathology according to BrainNet Europe guidelines52 and short post-mortem delay (Supplementary Table 1). Clinical parkinsonism during life fulfilling the UK Parkinson’s Disease Society Brain Bank53 or Movement Disorders Society54 criteria was verified through review of medical records. The selected brain autopsies were kindly provided by the Netherlands Brain Bank (www.brainbank.nl) and Normal Aging Brain Collection Amsterdam. The samples comprised 23 neuropathologically healthy individuals and 61 individuals with neuropathological changes corresponding to different Braak Lewy body stages. DNA and total RNA were simultaneously isolated from each sample. This was followed by genotyping and RNA sequencing (Supplementary material). Twenty-three neuropathologically healthy individuals at Braak Lewy body stage 0 (nMale = 12, nFemale = 11) and 19 individuals at Braak Lewy body stage 5 (nMale = 12, nFemale = 7) were included in the differential gene expression analysis. We chose to include samples from patients at Braak Lewy body stage 5 as this is the stage at which Lewy bodies first appear in the frontal cortex and is therefore likely to be subjected to more pronounced disease relevant changes in gene expression. Eighty-three samples were included in the eQTL analysis.
Differential gene expression analysis
The NEBNext Ultra RNA Library Prep Kit for Illumina was used for library preparation after removal of cytoplasmic and mitochondrial ribosomal RNAs. The library was paired-end sequenced on the NovaSeq 6000 platform and quality control was performed with FastQC55 and MultiQC.56 Salmon57 was used for alignment and quantification (see Supplementary material for details).
Differential gene expression between males and females was assessed using the DESeq2 R package (version 1.34.0).58 This was performed separately for the neuropathologically healthy group (Braak Lewy body stage 0, nMale = 12, nFemale = 11) and the disease group (Lewy body Braak stage 5, nMale = 12, nFemale = 7) (Supplementary Fig. 1). Mitochondrial genes were excluded and only genes with counts >0 in all samples (21 615 genes) were used in the analysis. Cell-type proportions for each sample were estimated using Scaden version 1.1.259 (Supplementary material). The quality surrogate variable analysis (qSVA) framework60 was used to estimate and remove RNA quality confounding in differential expression analysis (Supplementary material). Gene expression analysis was performed controlling for covariates (sex, age at death and two or four qSVs for Braak Lewy body Stage 0 and Braak Lewy body Stage 5, respectively) using the Wald test followed by FDR calculation using the Benjamini–Hochberg procedure. A Pearson’s chi-square test was used to determine whether there was a statistically significant enrichment of genes located near the Parkinson’s disease loci among the differentially expressed genes between males and females in the Braak Lewy body Stage 0.
Sex-specific expression quantitative trait locus analyses
Gene expression counts for 83 samples were transformed with vst() function from DESeq258 and adjusted for age at death, Braak Lewy body stage and five qSVs. Count correction was performed using the removeBatchEffect() function from the R package limma (3.50.0).61The list of 90 Parkinson’s disease risk loci identified in the largest Parkinson’s disease GWAS19 was used to extract the nearest gene and QTL nominated genes to each SNP. The association between the Parkinson’s disease variants and the expression of the nearest gene and QTL nominated genes to each SNP was investigated. Moreover, the association between the variants located in the 11 pleiotropy loci identified in the conjunctional analysis and their mapped genes was investigated. Genes not expressed in our samples and HLA genes were excluded from the analysis. The association between the adjusted expression of 87 GWAS genes and 91 pleiotropy genes and the genotype of the corresponding SNP were tested separately for each sex (nMale = 41, nFemale = 42) using a linear regression model [lm() function in R]. For the significant eQTLs in either males (GWAS = 7, pleiotropy = 5) or females (GWAS = 6, pleiotropy = 1), we fitted a linear model (Adjusted counts of gene of interest ∼ Genotype × Sex) using the lm() function and compared the slopes using pairs() function in R to assess whether the genotype had the same effect on gene expression in both males and females or not. Since the analyses included only a small selection of genes mapped from genome-wide significant SNPs and tested only against the corresponding SNP, a nominal P-value was used as the threshold of significance (P-value < 0.05).
Age-related expression analyses
We first compared the influence that ageing has on the adjusted counts of the genes located in the 90 Parkinson’s disease risk loci (87 genes)19 and in the 11 identified pleiotropy loci (91 genes) by fitting a linear model (adjusted counts of gene of interest ∼ Age × Sex). Then, we compared the slopes of the fitted models using pairs() function in R to determine whether or not ageing had the same effect in both sexes. For the genes that were affected by age in a sex-specific manner (nine GWAS genes and nine pleiotropy genes), we investigated the association between the ageing effect on gene expression and the genotype of the nearby SNP separately in females and males by fitting a linear model (adjusted counts of gene of interest ∼ Age × Genotype) and then compared the slopes of the models fitted to each genotype using pairs() function in R. The lm() function in R was used to fit the linear models. The analyses included only a small selection of genes mapped from genome-wide significant SNPs, therefore, a nominal P-value was used as the threshold of significance (P-value < 0.05) for these analyses.
Ethics
For the data from the GWASs, information about patient consents and ethical considerations are described in the original publications. Written informed consent was collected from the brain donors or their next of kin for the use of clinical information and tissue samples for research purposes. The gene expression part of this study was approved The Regional Committee for Medical and Health Research Ethics South-East Norway and the Medical Ethics Committee of the VU University Medical Centre, Amsterdam.
Results
Genetic overlap and correlation between Parkinson’s disease and sex-specific traits
To study the genetic component to sex differences in Parkinson’s disease, we investigated polygenic overlap between Parkinson’s disease19 and the sex-specific traits age at menarche20 and age at menopause.21 The reversed stratified and stratified quantile-quantile plots (Q-Q plots) indicate successive increments of SNP enrichment for Parkinson’s disease conditioned on association P-values for the sex-specific traits age at menarche and age at menopause (Fig. 1A) and the sex-specific traits conditioned on association P-values for Parkinson’s disease (Fig. 1B). This suggests polygenic overlap between the phenotypes. At conditional FDR <0.05, we identified 215 loci associated with age at menarche and 148 loci associated with age at menopause conditional on their association with Parkinson’s disease (Supplementary Tables 2 and 3).
Gene loci shared between sex-specific traits and Parkinson’s disease
Performing conjunctional FDR analysis (FDR < 0.05), we identified seven loci shared between Parkinson’s disease and age at menarche (Supplementary Table 4) and four loci shared between Parkinson’s disease and age at menopause (Supplementary Table 5). The chromosomal distribution of loci jointly associated with Parkinson’s disease and sex-specific traits are illustrated in Manhattan plots (Fig. 2). Eight of the 11 loci are novel for Parkinson’s disease, and six are novel for the sex-specific traits (Table 1).
Table 1.
Shared loci between Parkinson’s disease and sex-specific traits
| Sex-specific trait | Chr | SNP | Novelty | eQTL mapping | Gene expression in brain | |
|---|---|---|---|---|---|---|
| Sex-specific eQTL | Sex-specific age-related expression | |||||
| Menarche | 1 | rs708723 | In menarche | ELK4, SLC45A3, NUCKS1, RAB7L1, SLC41A1, PM20D1, SLC26A9, AVPR1B, C1orf186, CTSE | – | NUCKS1, SLC45A3 |
| Menarche | 1 | rs1352162 | In Parkinson’s disease | CEP170, SDCCAG8, AKT3, ZBTB18, C1orf100, ADSS, C1orf101, EFCAB2 | – | ADSS, EFCAB2 |
| Menopause | 2 | rs780104 | In Parkinson’s disease | DPYSL5, TMEM214, OST4, EMILIN1, KHK, CGREF1, ABHD1, PREB, SLC5A6, ATRAID, CAD, TRIM54, MPV17, GTF3C2, EIF2B4, SNX17, ZNF513, PPM1G, NRBP1, KRTCAP3, IFT172, FNDC4, GCKR, ZNF512, CCDC121, GPN1, SUPT7L, SLC4A1AP, PPP1CB | – | OST4, PREB, ZNF513, IFT172 |
| Menopause | 3 | rs905604 | In all | NIT2, TOMM70A, LNP1, TFG, ABI3BP | – | – |
| Menarche | 3 | rs843351 | In Parkinson’s disease | MCCC1, ABCC5, EIF2B5, DVL3, AP2M1, ABCF3, VWA5B2, ECE2, CAMK2N2, EIF4G1 | – | – |
| Menarche | 6 | rs660895 | In menarche | BAG6, LY6G5B, LY6G6E, LY6G6D, C6orf25, LY6G6C, DXO, STK19, C4A, C6orf10, HLA-DRB1, -DQA1, -DQB1, -DQA2, -DQB2, -DOB, TAP2 | – | – |
| Menopause | 6 | rs6932585 | In menopause | CEP85L, MCM9, ASF1A | – | – |
| Menarche | 10 | rs12571664 | In Parkinson’s disease | TIAL1, INPP5F, MCMBP, SEC23IP | MCMBP | – |
| Menarche | 12 | rs10843831 | In all | ERGIC2, OVCH1-AS1, OVCH1, IPO8, CAPRIN2, TSPAN11 | ERGIC2 | – |
| Menopause | 12 | rs7953894 | In Parkinson’s disease | ABCB9, SBNO1, SETD8, RILPL2, DDX55, ATP6V0A2 | – | – |
| Menarche | 17 | rs178830 | In all | ADORA2B, ZSWIM7, TTC19, NCOR1, PIGL, CENPV, UBB, TRPV2, CCDC144A | – | CCDC144A |
Chr = chromosome; eQTL = expression quantitative trait loci; SNP = single nucleotide polymorphism.
We mapped 64 and 43 protein-coding genes to candidate SNPs jointly associated with Parkinson’s disease and each of age at menarche and age at menopause, respectively (Supplementary Tables 6 and 7). To do that, we employed three strategies: positional mapping, eQTL and chromatin mapping. We performed gene-set analyses in FUMA40 on each of these groups of genes. While there were no significantly enriched terms for the genes mapped to the shared loci between Parkinson’s disease and age at menopause gene-sets, 59 gene-sets were enriched with mapped genes for Parkinson’s disease and age at menarche. Among these gene-sets there was a predominance of gene-sets related to immune function (e.g. ‘antigen processing and presentation’ and ‘positive regulation of immune response’) and vesicle trafficking (e.g. ‘clathrin coated endocytic vesicle’ and ‘endoplasmic reticulum to Golgi transport vesicle membrane’) (Supplementary Table 8). Moreover, pathway analyses revealed a predominance of pathways related to immune function (e.g. ‘MHC class II antigen presentation’, ‘antigen processing and presentation’ and ‘phagosome’), infection control (‘viral myocarditis’ and ‘staphylococcus aureus infection’) and autoimmunity (‘asthma’ and ‘type I diabetes mellitus’) from genes jointly associated with Parkinson’s disease and age a menarche (Supplementary Table 9).
We also present spatiotemporal gene-expression analyses of mapped genes for Parkinson’s disease and age at menarche using normalized BrainSpan RNA sequencing data (Fig. 3), showing highest gene expression of genes jointly associated with Parkinson’s disease and age at menarche directly prior to expected menarche.
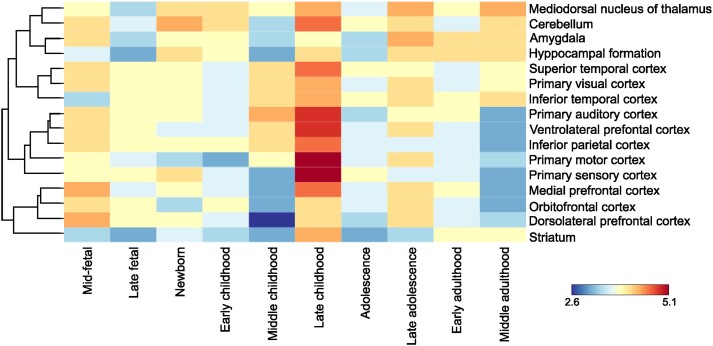
Figure 3.
Spatiotemporal gene expression of all mapped genes to shared loci between Parkinson’s disease and age at menarche. The dendrogram and heat map were built using RNA sequencing data from BrainSpan over 11 developmental periods (columns) and 16 brain regions (rows). Gene expression was measured as log2 transformed RPKM (reads per kilobase per million mapped reads). Global expression of mapped genes to shared loci between Parkinson’s disease and age at menarche had the highest expression during late childhood, most prominent in neocortical areas. Gene expression varied from a minimum of 2.6 log2(RPKM) and to a maximum of 5.1 log2(RPKM).
Sex-specific gene expression differences in healthy individuals and Parkinson’s disease patients
We investigated sex-specific gene expression in the frontal cortex of neuropathologically healthy individuals (Braak Lewy body Stage 0) and of Parkinson’s disease patients (Braak Lewy body Stage 5). We found a total of 142 differentially expressed genes (FDR < 0.05) in the neuropathologically healthy donors (Supplementary Table 10), whereas when looking at differences in gene expression between male and female donors at Braak Lewy body Stage 5 we found only 11 differentially expressed genes (FDR < 0.05) (Supplementary Table 11). None of the differentially expressed genes at Braak Lewy body Stage 5 were among the genes differentially expressed in the neuropathologically healthy group.
None of the genes mapped to the identified shared loci between Parkinson’s disease and sex-specific traits was among the genes differentially expressed between males and females either in the neuropathologically healthy individuals or the Parkinson’s disease patients.
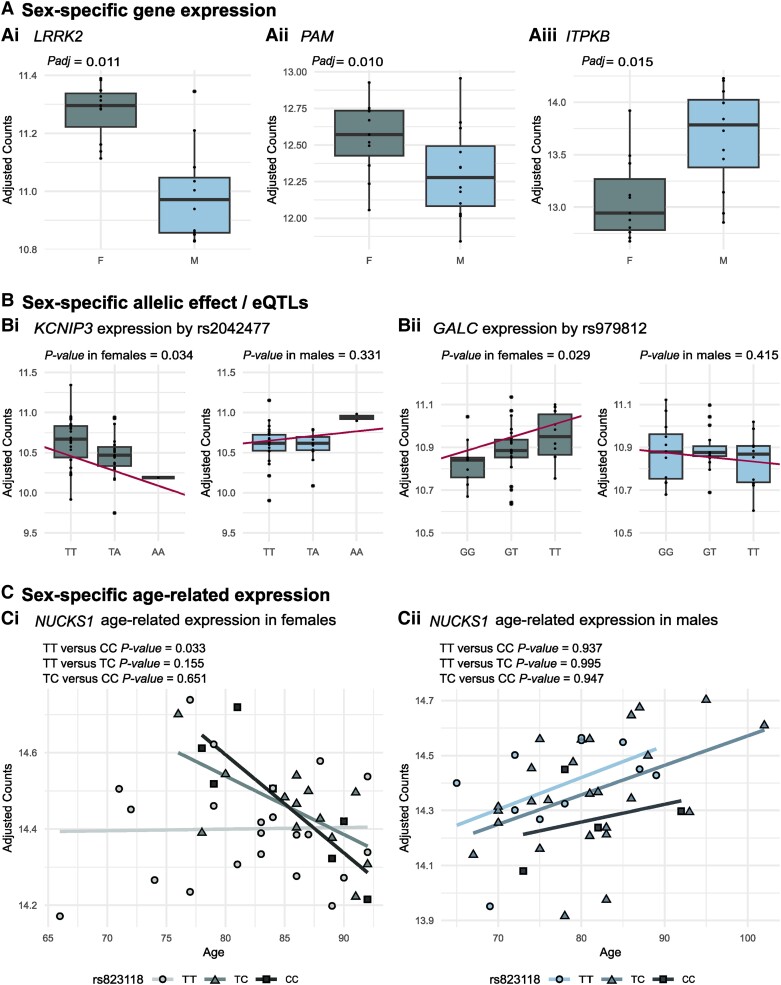
Three genes that are the nearest genes to significant lead Parkinson’s disease GWAS19 SNPs, LRRK2 (rs76904798 and rs34637584), PAM (rs26431) and ITPKB (rs4653767) were among the genes differently expressed in the neuropathologically healthy group between males and females (Fig. 4A). LRRK2 and PAM were upregulated in female samples as compared to male samples, whereas ITPKB was downregulated. There was a statistically significant enrichment of Parkinson’s disease risk genes among the genes that were differentially expressed between males and females in the neuropathologically healthy group (P = 0.004).
Figure 4.
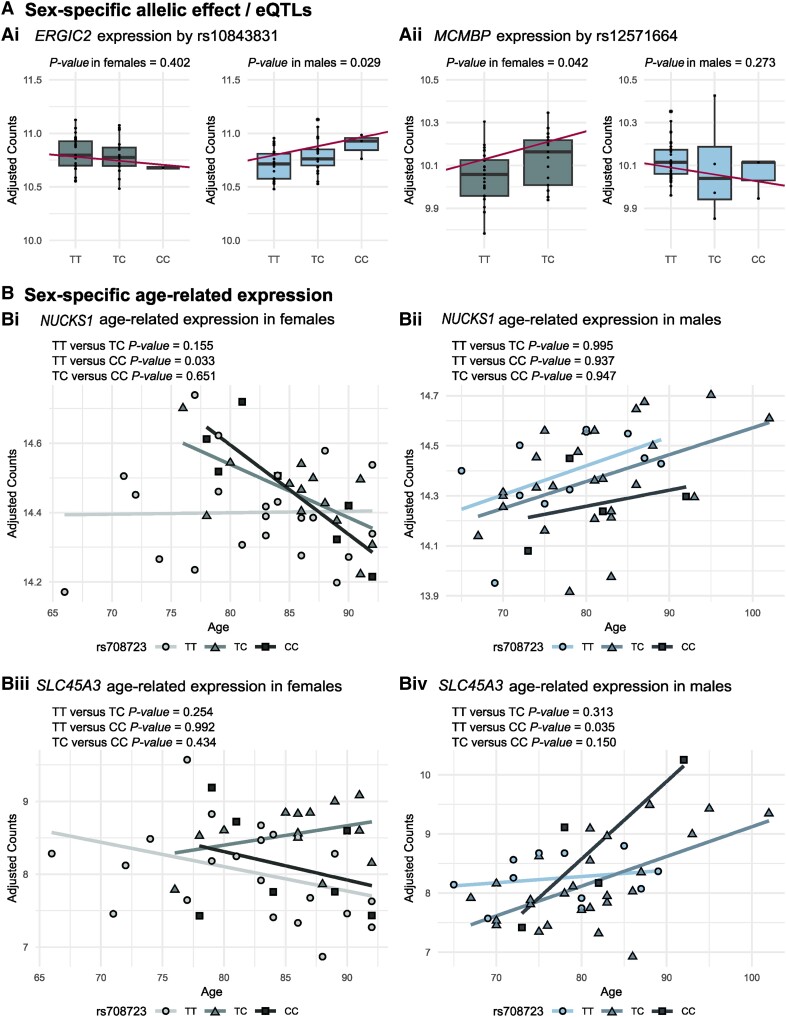
Sex-specific expression of genes mapped to Parkinson’s disease loci. (A) Sex-specific expression of genes mapped to Parkinson’s disease loci in the superior frontal cortex of neuropathologically healthy donors: [A(i)] LRRK2 (rs76904798 and rs34637584) and [A(ii)] PAM (rs26431) show higher expression levels in females (F) compared to males (M). [A(iii)] ITPKB (rs4653767) shows lower expression levels in females (F) compared to males (M). (B) Genes showing a sex-specific allelic effect/expression quantitative trait loci (eQTL) and a statistically different direction in the two sexes: [B(i)] KCNIP3 (rs2042477) and [B(ii)] GALC (rs979812). (C) Of the nine genes with statistically significant different age-related expression between the sexes, NUCKS1 sex-specific age-related expression is associated with the rs823118 genotype in [C(i)] females but not [C(ii)] males. A = adenine; C = cytosine; G = guanine; T = thymine.
Of note, no statistically significant differences in cell composition were found between males and females either in the neuropathologically healthy group or in the Braak Lewy body Stage 5 group after correcting for multiple testing for all the cell-types (Supplementary material and Supplementary Fig. 2).
Sex-specific eQTL of candidate genes
We investigated whether the identified pleiotropy and Parkinson’s disease SNPs could have a sex-specific effect on gene expression that could contribute to the increased disease risk observed in the male population.
Out of 101 genes (excluding six HLA genes) mapped to the 11 shared loci between Parkinson’s disease and the sex-specific traits, 91 were expressed in our samples. A total of six genes from four loci were nominal eQTLs exclusively in one sex (five in males and one in females) (Supplementary Table 12 and Supplementary Fig. 3). Moreover, two of these genes (rs10843831, ERGIC2; and rs12571664, MCMBP) showed a different direction of the change in expression in the two sexes (P < 0.05) (Fig. 5A).
Figure 5.
Sex-specific expression of genes mapped to loci shared between Parkinson’s disease and sex-specific traits. (A) Genes showing a sex-specific allelic effect/expression quantitative trait loci (eQTL) and a statistically different direction in the two sexes: [A(i)] ERGIC2 (rs10843831) and [B(ii)] MCMBP (rs12571664). (B) Of the nine genes with a statistically significant different age-related expression between the sexes, [B(i and ii)] NUCKS1 and [B(iii and iv)] SLC45A3 sex-specific age-related expression is associated with rs708723 genotype (Locus 1 of loci shared between Parkinson’s disease and age at menarche) exclusively in female or male samples, respectively. C = cytosine; T = thymine.
Furthermore, we found that 87 of the 97 Parkinson’s disease genes19 were expressed in our samples. CAB39L was a nominal eQTL both in the male and female samples with the risk allele T being associated with an increase in the expression levels in both sexes (Supplementary Fig. 4J). Six genes were nominal eQTLs exclusively in the male samples and five genes were nominal eQTLs exclusively in the female samples (Supplementary Table 13 and Supplementary Fig. 4). Only 2 of these 11 eQTLs (rs2042477, KCNIP3; and rs12571664, GALC) showed a different direction of the change in expression in the other sex (P < 0.05) (Fig. 4B).
Candidate genes show age-related sex-specific expression
As ageing is known to influence gene expression in a sex-specific manner18 and is the main risk factor for Parkinson’s disease,62 we next investigated age-related gene expression differences between males and females for the genes located in the 11 shared loci between Parkinson’s disease and sex-specific traits and in Parkinson’s disease risk loci.
We found that the age-related expression of nine genes from three loci shared between Parkinson’s disease and sex-specific traits (NUCKS1, SLC45A3, ADSS, EFCAB2, CCDC144A, IFT172, OST4, PREB and ZNF513) was nominally statistically significantly different between males and females (Supplementary Table 14 and Supplementary Fig. 5). Next, we focused on these genes showing sex-specific age-related expression and checked whether the genotype of the SNPs located in these loci was influencing age-related gene expression in a sex-specific manner. We found that the age-related expression of two genes (NUCKS1 and SLC45A3) located in the menarche locus 1 was also associated with the genotypes of a SNP (rs708723) exclusively in female or in male samples, respectively (Fig. 5B).
Moreover, we found that the age-related expression of nine Parkinson’s disease genes (NUCKS1, SIPA1L2, STK39, SCARB2, CLCN3, RNF141, GALC, DNAH17 and DCAF16) was statistically significantly different between males and females (Supplementary Table 15 and Supplementary Fig. 6). Next, we focused on these genes showing sex-specific age-related expression and checked whether the genotype of the SNPs located in each locus was influencing the age-related gene expression in a sex-specific manner. We found that rs823118 influences the age-dependent expression of NUCKS1 only in females (Fig. 4C).
Discussion
Here, by combining large GWAS datasets from Parkinson’s disease and sex-specific traits with in-depth RNA sequencing data from human brains, we investigated sex-specific differences associated with Parkinson’s disease that could partly explain the increased incidence of the disease in the male population. We thereby demonstrate that the lack of clear sex differences in allele frequencies for Parkinson’s disease SNPs6-8 does not exclude a genetic contribution to sex differences in incidence.
By performing conjunctional FDR analyses, the present study revealed that 11 genomic loci are jointly associated with Parkinson’s disease and sex-specific traits. Intriguingly, only three of these loci had been previously associated with Parkinson’s disease and none is directly linked to oestrogen signalling pathways. However, some of the genes mapped to the identified SNPs are expressed in the ovaries (Supplementary Table 6), and therefore their expression could still be mediated by oestrogen signalling pathways. The mechanisms through which oestrogens might reduce the risk of Parkinson’s disease are not fully understood. It is possible that the SNPs located in these genomic loci could for example interact with oestrogen receptors that are widespread in the brain22 or with other related pathways such as the phosphatidylinositol 3 kinase (PI3K)/Akt pathway26 and the extracellular signal-regulated kinase (ERK1/2) pathway,27 ultimately contributing to the observed sex-specific differences in Parkinson’s disease. Post-translational modulations, such as the phosphorylation that inhibits the pro-apoptotic protein Bad, are proven important in Parkinson’s disease. Moreover, oestrogens are known to affect gene expression directly or indirectly.
We mapped genes to the 11 genomic loci associated with Parkinson’s disease and sex-specific traits. These genes were used to perform gene-set and pathway analyses. A large portion of the gene-sets enriched with genes mapped to loci shared between Parkinson’s disease and age at menarche are related to processes known to be important for Parkinson’s disease.63 These include activation and regulation of the immune response and gene-sets that belong to cellular compartments involved in endosomes and MHC process. Pathway analysis further highlighted the importance of the immune system as it identified pathways related to autoimmune diseases, infectious diseases, antibody production and antigen presentation. These results revealed putative mechanisms important for the sex differences in Parkinson’s disease, like sex differences in immune responses64,65 and even sex differences in microglial phenotypes.66,67
The spatiotemporal gene-expression analysis identified developmental time periods where these mechanisms set to action. Interestingly, genes mapped from loci shared between Parkinson’s disease and age at menarche are expressed in the human brain at the highest levels in late childhood, fitting with the years prior to menarche, and highlighting how processes early in life might affect diseases that tend to develop late in life. The fact that gene expression during this time period was highest in cortical structures is particularly intriguing, since cortical structures are known to be affected by the neuropathological hallmarks of Parkinson’s disease only in late stages of the disease.68 Spatiotemporal gene-expression analysis was only performed with genes mapped to loci shared between Parkinson’s disease and age at menarche, as spatiotemporal mapping was not available for older age-groups than middle adulthood and hence did not cover the age-range where menopause tends to occur.
We did not find significant gene-sets or pathways for the genes mapped to loci overlapping between Parkinson’s disease and age at menopause. The primary reason for stronger genetic overlap between Parkinson’s disease and age at menarche than age at menopause is likely to be differences in sample size. Moreover, the exact age at menopause, defined as 12 months of amenorrhoea, would likely be more approximate than age at menarche, due to a variety of symptoms ranging from hot flashes and night sweats to irregular periods when approaching menopause. So, in addition to the higher statistical power, data on age at menarche is likely to be more accurate than the age at menopause.
To highlight genes that might be involved in the mechanisms leading to an increased disease incidence in the male population and that consequentially should be further prioritized for experimental validation, we performed sex-specific, age-related gene expression analysis and sex-specific eQTL analysis.
We found that differences in gene expression between males and females are more prominent in the neuropathologically healthy donors than in patients with Parkinson’s disease and that there is no overlap between the two groups. This might be because the transcriptome of females that develop Parkinson’s disease is more similar to the male transcriptome, compared to females that stay neurologically healthy throughout their life. Further, disease might affect gene expression to an extent that masks sex differences.
None of the genes mapped to the shared loci between Parkinson’s disease and sex-specific traits was among the differentially expressed genes. However, we found an enrichment of genes located in known Parkinson’s disease risk loci identified in GWAS19 (LRRK2, PAM and ITPKB) among the genes differentially expressed between the sexes in the neuropathologically healthy individuals. LRRK2 mutations comprise the most common cause of the autosomal dominant form of Parkinson’s disease69 and genetic variation in the LRRK2 locus is associated with an increased risk of sporadic Parkinson’s disease. LRRK2 encodes leucine-rich repeat kinase 2, which phosphorylates a broad range of proteins involved in multiple processes such as neuronal plasticity, autophagy, vesicle trafficking and dopaminergic neuron apoptosis.70-73 In our study, we found that LRRK2 could also play a role in increasing Parkinson’s disease incidence in males. No studies, other than the Parkinson’s disease GWAS,19 have linked PAM to Parkinson’s disease. However, PAM is highly expressed in the rat uterus and regulated by oestrogens.74 We further found higher PAM expression levels in females; therefore, also in human brain, oestrogen levels might regulate PAM expression. However, further investigations are needed to understand the possibly protective role that this gene has in disease development and how it might influence disease incidence. ITPKB, in addition to the association with Parkinson’s disease GWAS SNP rs4653767, has been linked to Alzheimer’s disease where it has been shown to have higher expression levels in the cerebral cortex of patients than in control subject and to exacerbate Alzheimer’s pathology in mice as it is involved in the regulation of neuronal cell apoptosis.75 Moreover, ITPKB has been involved in Alzheimer’s disease pathogenesis in females.76 The ITPKB upregulation that we observed in males could therefore play a similar role in Parkinson’s disease patients, not observed in females.
Since the existence of sex-specific eQTLs has been demonstrated,17 we performed sex-specific eQTL analyses for the newly identified shared loci between Parkinson’s disease and the sex-specific traits and for the 90 Parkinson’s disease risk SNPs.19 For these analyses, we set a threshold for significance at a nominal P-value of 0.05. To strengthen the validity of our results, we further investigated the significant eQTLs by comparing the effect that the genotype has on the direction of the expression in females and males. We suggest that the expression of two genes located in the 11 shared loci between Parkinson’s disease and sex-specific traits may be influenced in a sex-specific manner by the nearby SNP, showing a different effect in the two sexes. The rs10843831 SNP, associated with age at menarche, was a putative sex-specific eQTL for the ERGIC2 gene as an increase in its expression was associated with the C allele only in males, and the opposite trend was observed in females [Fig. 5A(i)] ERGIC2 encodes the endoplasmic reticulum-Golgi intermediate compartment protein 2. Little is known about this protein but based on its sequence and structure similarity to the protein encoded by ERGIC1, it has been suggested that it has a possible role in transport between the endoplasmic reticulum and Golgi.77 Because of the fundamental role that the endoplasmic reticulum plays in regulating protein homeostasis, this organelle has been implicated with Parkinson’s disease.78 In females, the rs12571664 SNP, associated with age at menarche, was a putative sex-specific eQTL for the MCMBP gene showing the opposite trend in males [Fig. 5A(ii)]. MCMBP encodes the mini-chromosome maintenance complex-binding proteins acting as a regulator of DNA replication.79 Further studies are needed to elucidate whether these genes play a role in Parkinson’s disease development and increased incidence in the male population.
When investigating Parkinson’s disease SNPs, we found that two of them influenced the expression of KCNIP3 and GALC in a sex-specific manner showing also a statistically significant different effect in the two sexes. However, little is known about the functions of the proteins encoded by these genes and further studies are needed to elucidate whether they have a role in Parkinson’s disease and, if so, how they could affect disease risk in a sex-specific manner.
Finally, as age is the main risk factor for Parkinson’s disease and age-related gene expression can be sex-specific,18 we explored the hypothesis that the identified shared SNPs between Parkinson’s disease and sex-specific traits and the Parkinson’s disease risk SNPs could be associated with changes in age-related gene expression in a sex-specific manner. We first identified the genes showing sex-specific age-related expression setting a threshold for significance at a nominal P-value of 0.05; next, we investigated whether the age-related sex-specific expression of the significant genes was also influenced by the genotype. Only two of the genes, SLC45A3 and NUCKS1, were nominally significant in both levels of analyses. The age-related sex-specific expression of both these genes was associated with the rs708723 SNP shared between Parkinson’s disease and age at menarche. Interestingly, the age-related sex-specific expression of NUCKS1 was also associated with the Parkinson’s disease SNP rs823118. Therefore, even though these results were only nominally significant, we suggest that this locus, and in particular of NUCKS1, could play an important role in Parkinson’s development and that it could be linked to sex-specific differences of the disease.
Together, these results demonstrated the strength of the pleiotropy analysis. However, we acknowledge that there were some limitations to the current study. First, the Parkinson’s disease GWAS19 included sex as a covariate possibly affecting the ability to identify sex-specific effects. Nonetheless, it has been shown that there is a high genetic correlation between the male and female Parkinson’s disease GWAS7 and that there are no sex differences in Parkinson’s disease risk alleles neither in autosomes7 nor sex chromosomes.6,8 For these reasons, we believe that our approach of identifying loci that are pleiotropic with both Parkinson’s disease and sex-specific age-related traits allowed us to capture biologically relevant gene loci, which were further verified using gene expression data from the human brain. Additionally, the sex-specific traits used in this study focused on the length of the reproductive life in females, without including factors that might temporarily increase (e.g. pregnancies) or decrease (e.g. breastfeeding) the circulating oestrogen levels. Also, while sex hormone levels rise fast around age at menarche, the decrease is more gradual starting already from the end of the third decade of life, until most females enter menopause in their fifties. Only at this point do the oestrogen levels in females become more comparable to the levels in males. Furthermore, while functionally annotating candidate SNPs reduces the probability of missing causal variants, this approach increases the number of false positives to the gene-mapping, gene-set enrichment and gene-expression analyses. We also acknowledge the small samples size and consequently the low statistical power of the RNA sequencing data used. However, we think that these limitations were counterbalanced by the stringent criteria used to select the samples included and by the careful selection of covariates which allowed us to exclude possible difference in gene expression caused by difference in cell composition or RNA quality between the analysed groups. Moreover, we acknowledge that setting a nominal P-value of 0.05 as threshold for significance for the eQTL and age-related expression analysis might lead to false positive findings. However, the analysis that employed this P-value included only a small number of genes and loci already found to be significant in other genome-wide analyses that adjusted for multiple testing. Additionally, to limit the number of false positives identified in the sex-specific eQTL analysis, we further prioritized only the eQTLs showing a significantly different change in expression in the two sexes; whereas, in the age-related sex-specific expression analysis, we highlighted only the genes whose age-related expression was also influenced by the corresponding SNP. Therefore, we believe that by combining several layers of analysis we were able to highlight genes likely involved in the disease and in the mechanisms leading to an increased incidence in the male population. However, further functional studies are needed to confirm our findings.
We also note that although our horizontal pleiotropy approach identifies genetic overlap between Parkinson’s disease and oestrogen-related traits, these results do not determine whether a causal link between hormone levels and Parkinson’s disease risk is driving this signal. Further research will be needed to disentangle the causal mechanisms, where Mendelian randomization could be a relevant genetic method.80
Not only Parkinson’s disease, but several neurological diseases, show differences in incidence between sexes. Examples of this being higher frequencies of Alzheimer’s disease81 and migraine82 in females. Our approach, using conjunctional FDR analyses to reveal pleiotropy between Parkinson’s disease and sex-specific traits, holds the potential to be valuable also for other neurological diseases.
In conclusion, in our study we identified new variants which show pleiotropy between Parkinson’s disease and age at menarche or age at menopause. This demonstrates a genetic contribution to differences in Parkinson’s disease incidence, despite the lack of sex differences in allele frequencies. These findings have implications for the understanding of how sex affect gene function and expression, and conjunctional FDR analysis could hence be used to detect functional genetic variance even in cases with no differences in allele frequency. Moreover, gene expression and eQTL analysis point to candidate genes for future studies that could possibly explain the higher incidence of Parkinson’s disease in males supporting the existence of sex-biased eQTLs for Parkinson’s disease risk variants. This shared genetic pattern between Parkinson’s disease and sex-specific traits underlines the importance of continued efforts to understand the biological processes that explain the lower Parkinson’s disease incidence in females, as potentially protective biological processes could be future targets for disease modifying drugs.
Supplementary Material
Acknowledgements
The work was performed on the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT Department (USIT) and on resources provided by UNINETT Sigma2—the National Infrastructure for High Performance Computing and Data Storage in Norway. The age at menarche GWAS summary statistics used in this study were generated including data from 23andMe, Inc. We would like to thank the research participants and employees of 23andMe for making this work possible. The authors are grateful to the Netherlands Brain Bank and its funders for providing the samples that made this study possible.
Contributor Information
Kaja Nordengen, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway; Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, 0372 Oslo, Norway.
Chiara Cappelletti, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway; Department of Mechanical, Electronics and Chemical Engineering, Faculty of Technology, Art and Design, OsloMet—Oslo Metropolitan University, 0130 Oslo, Norway; Department of Research, Innovation and Education, Oslo University Hospital, 0424 Oslo, Norway.
Shahram Bahrami, Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, 0372 Oslo, Norway; Norwegian Centre for Mental Disorders Research (NORMENT), Division of Mental Health and Addiction, Oslo University Hospital, 0450 Oslo, Norway.
Oleksandr Frei, Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, 0372 Oslo, Norway; Norwegian Centre for Mental Disorders Research (NORMENT), Division of Mental Health and Addiction, Oslo University Hospital, 0450 Oslo, Norway.
Lasse Pihlstrøm, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway.
Sandra Pilar Henriksen, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway.
Hanneke Geut, Section of Clinical Neuroanatomy and Biobanking, Department of Anatomy and Neurosciences, Amsterdam UMC, Location Vrije Universiteit Amsterdam, Amsterdam Neuroscience, 1081 Amsterdam, The Netherlands.
Annemieke J M Rozemuller, Department of Pathology, Amsterdam UMC, Location Vrije Universiteit Amsterdam, Amsterdam Neuroscience, 1081 Amsterdam, The Netherlands.
Wilma D J van de Berg, Section of Clinical Neuroanatomy and Biobanking, Department of Anatomy and Neurosciences, Amsterdam UMC, Location Vrije Universiteit Amsterdam, Amsterdam Neuroscience, 1081 Amsterdam, The Netherlands.
Ole A Andreassen, Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, 0372 Oslo, Norway; Norwegian Centre for Mental Disorders Research (NORMENT), Division of Mental Health and Addiction, Oslo University Hospital, 0450 Oslo, Norway.
Mathias Toft, Department of Neurology, Oslo University Hospital, 0424 Oslo, Norway; Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, 0372 Oslo, Norway.
Data availability
Data supporting the findings of this study are openly available from an online repository or are available on request from study authors. RNA-sequencing data can be accessed through the Gene Expression Omnibus (accession ID: GSE216281). All code is freely available at https://github.com/chiaracapp/Gene-Expression-and-Genetic-Sex-Differences-in-PD.git Analyses were conducted in Python v.3.5, MATLAB R2020b and R v.4.2.2. Locus definition, functional annotation and gene-set analysis were performed using FUMA (https://fuma.ctglab.nl/).40
Funding
The authors were funded by the Research Council of Norway (O.A.A.: 213837, 223273, 248778, 273291, 262656, 229129, 283798, 311993), the South-Eastern Norway Regional Health Authority (O.A.A.: 2013- 123, 2017-112, 2019-108;), the Norwegian Health Association (K.N.: 25598, S.B.: 22731) and the Department of Neurology at Oslo University Hospital (K.N.).
Competing interests
O.A.A. has received speaker’s honoraria from Sunovion and Lundbeck and is a consultant for cortechs.ai. The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Poewe W, Seppi K, Tanner CM, et al. . Parkinson Disease. Nat Rev Dis Primers. 2017;3:17013. [DOI] [PubMed] [Google Scholar]
- 2. Moisan F, Kab S, Mohamed F, et al. . Parkinson Disease male-to-female ratios increase with age: French nationwide study and meta-analysis. J Neurol Neurosurg Psychiatry. 2016;87:952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taylor KS, Cook JA, Counsell CE. Heterogeneity in male to female risk for Parkinson's disease. J Neurol Neurosurg Psychiatry. 2007;78:905–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nandipati S, Litvan I. Environmental exposures and Parkinson’s disease. Int J Environ Res Public Health. 2016;13:881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jurado-Coronel JC, Cabezas R, Rodríguez MFÁ, Echeverria V, García-Segura LM, Barreto GE. Sex differences in Parkinson’s disease: Features on clinical symptoms, treatment outcome, sexual hormones and genetics. Front Neuroendocrinol. 2018;50:18–30. [DOI] [PubMed] [Google Scholar]
- 6. Le Guen Y, Napolioni V, Belloy ME, et al. . Common X-chromosome variants are associated with Parkinson disease risk. Ann Neurol. 2021;90:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blauwendraat C, Iwaki H, Makarious MB, et al. . Investigation of autosomal genetic sex differences in Parkinson's disease. Ann Neurol. 2021;90:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grenn FP, Makarious MB, Bandres-Ciga S, et al. . Analysis of Y chromosome haplogroups in Parkinson's disease. Brain Commun. 2022;4:fcac277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sawada H, Shimohama S. Estrogens and Parkinson disease: Novel approach for neuroprotection. Endocrine. 2003;21:77–79. [DOI] [PubMed] [Google Scholar]
- 10. Brann DW, Dhandapani K, Wakade C, Mahesh VB, Khan MM. Neurotrophic and neuroprotective actions of estrogen: basic mechanisms and clinical implications. Steroids. 2007;72:381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hirohata M, Ono K, Morinaga A, Ikeda T, Yamada M. Anti-aggregation and fibril-destabilizing effects of sex hormones on alpha-synuclein fibrils in vitro. Exp Neurol. 2009;217:434–439. [DOI] [PubMed] [Google Scholar]
- 12. Ragonese P, D'Amelio M, Salemi G, et al. . Risk of Parkinson disease in women: Effect of reproductive characteristics. Neurology. 2004;62:2010–2014. [DOI] [PubMed] [Google Scholar]
- 13. Canonico M, Pesce G, Bonaventure A, et al. . Increased risk of Parkinson's disease in women after bilateral oophorectomy. Mov Disord. 2021;36:1696–1700. [DOI] [PubMed] [Google Scholar]
- 14. Ragonese P, D'Amelio M, Callari G, Salemi G, Morgante L, Savettieri G. Age at menopause predicts age at onset of Parkinson's disease. Mov Disord. 2006;21:2211–2214. [DOI] [PubMed] [Google Scholar]
- 15. Oliva M, Munoz-Aguirre M, Kim-Hellmuth S, et al. . The impact of sex on gene expression across human tissues. Science. 2020;369:eaba3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lopes-Ramos CM, Chen CY, Kuijjer ML, et al. . Sex differences in gene expression and regulatory networks across 29 human tissues. Cell Rep. 2020;31:107795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trabzuni D, Ramasamy A, Imran S, et al. . Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun. 2013;4:2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berchtold NC, Cribbs DH, Coleman PD, et al. . Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nalls MA, Blauwendraat C, Vallerga CL, et al. . Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Day FR, Thompson DJ, Helgason H, et al. . Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet. 2017;49:834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Day FR, Ruth KS, Thompson DJ, et al. . Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47:1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brailoiu E, Dun SL, Brailoiu GC, et al. . Distribution and characterization of estrogen receptor G protein-coupled receptor 30 in the rat central nervous system. J Endocrinol. 2007;193:311–321. [DOI] [PubMed] [Google Scholar]
- 23. Morissette M, Le Saux M, D'Astous M, et al. . Contribution of estrogen receptors alpha and beta to the effects of estradiol in the brain. J Steroid Biochem Mol Biol. 2008;108(3–5):327–338. [DOI] [PubMed] [Google Scholar]
- 24. Al Sweidi S, Sanchez MG, Bourque M, Morissette M, Dluzen D, Di Paolo T. Oestrogen receptors and signalling pathways: Implications for neuroprotective effects of sex steroids in Parkinson's disease. J Neuroendocrinol. 2012;24:48–61. [DOI] [PubMed] [Google Scholar]
- 25. Bourque M, Dluzen DE, Di Paolo T. Signaling pathways mediating the neuroprotective effects of sex steroids and SERMs in Parkinson's disease. Front Neuroendocrinol. 2012;33:169–178. [DOI] [PubMed] [Google Scholar]
- 26. Quesada A, Lee BY, Micevych PE. PI3 Kinase/akt activation mediates estrogen and IGF-1 nigral DA neuronal neuroprotection against a unilateral rat model of Parkinson's disease. Dev Neurobiol. 2008;68:632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marino M, Galluzzo P, Ascenzi P. Estrogen signaling multiple pathways to impact gene transcription. Curr Genomics. 2006;7:497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu Y, Yang GY, Ahlemeyer B, et al. . Transforming growth factor-beta 1 increases bad phosphorylation and protects neurons against damage. J Neurosci. 2002;22:3898–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kochmanski J, Kuhn NC, Bernstein AI. Parkinson's disease-associated, sex-specific changes in DNA methylation at PARK7 (DJ-1), SLC17A6 (VGLUT2), PTPRN2 (IA-2beta), and NR4A2 (NURR1) in cortical neurons. NPJ Parkinsons Dis. 2022;8:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carroll JS, Meyer CA, Song J, et al. . Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. [DOI] [PubMed] [Google Scholar]
- 31. Parcellier A, Tintignac LA, Zhuravleva E, Hemmings BA. PKB And the mitochondria: AKTing on apoptosis. Cell Signal. 2008;20:21–30. [DOI] [PubMed] [Google Scholar]
- 32. Hetman M, Gozdz A. Role of extracellular signal regulated kinases 1 and 2 in neuronal survival. Eur J Biochem. 2004;271:2050–2055. [DOI] [PubMed] [Google Scholar]
- 33. Shi J, Wu L, Li B, et al. . Transcriptome-Wide association study identifies susceptibility loci and genes for age at natural menopause. Reprod Sci. 2019;26:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andreassen OA, Djurovic S, Thompson WK, et al. . Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smeland OB, Bahrami S, Frei O, et al. . Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. 2020;25:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smeland OB, Frei O, Shadrin A, et al. . Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet. 2020;139:85–94. [DOI] [PubMed] [Google Scholar]
- 37. Bahrami S, Nordengen K, Shadrin AA, et al. . Distributed genetic architecture across the hippocampal formation implies common neuropathology across brain disorders. Nat Commun. 2022;13:3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elvsashagen T, Shadrin A, Frei O, et al. . The genetic architecture of the human thalamus and its overlap with ten common brain disorders. Nat Commun. 2021;12:2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng W, Frei O, van der Meer D, et al. . Genetic association between schizophrenia and cortical brain surface area and thickness. JAMA Psychiatry. 2021;78:1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boyle AP, Hong EL, Hariharan M, et al. . Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roadmap Epigenomics C, Kundaje A, Meuleman W, et al. . Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu Z, Zhang F, Hu H, et al. . Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. [DOI] [PubMed] [Google Scholar]
- 45. Consortium GT, Laboratory DA. Coordinating center -analysis working G, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gene Ontology C . The gene ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ashburner M, Ball CA, Blake JA, et al. . Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11:1889–1907. [DOI] [PubMed] [Google Scholar]
- 49. Miller JA, Ding SL, Sunkin SM, et al. . Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bahl E, Koomar T, Michaelson JJ. Cerebroviz: An R package for anatomical visualization of spatiotemporal brain data. Bioinformatics. 2017;33:762–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.BrainSpan. BrainSpan atlas of the developing human brain. Accessed 16 October 2022. http://www.brainspan.org/
- 52. Alafuzoff I, Ince PG, Arzberger T, et al. . Staging/typing of Lewy body related alpha-synuclein pathology: A study of the BrainNet Europe Consortium. Acta Neuropathol. 2009;117:635–652. [DOI] [PubMed] [Google Scholar]
- 53. Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson's disease. J Neurol Neurosurg Psychiatry. 1988;51:745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Postuma RB, Berg D, Stern M, et al. . MDS Clinical diagnostic criteria for Parkinson's disease. Mov Disord. 2015;30:1591–1601. [DOI] [PubMed] [Google Scholar]
- 55. Andrews S. FastQC: A quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute; 2010. [Google Scholar]
- 56. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-Seq: Transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jaffe AE, Tao R, Norris AL, et al. . qSVA framework for RNA quality correction in differential expression analysis. Proc Natl Acad Sci U S A. 2017;114:7130–7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smyth GK, Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S. limma: Linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds. Bioinformatics and computational biology solutions using R and Bioconductor. Statistics for biology and health. Springer; 2005:397–420. [Google Scholar]
- 62. Reeve A, Simcox E, Turnbull D. Ageing and Parkinson's disease: Why is advancing age the biggest risk factor? Ageing Res Rev. 2014;14:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tan EK, Chao YX, West A, Chan LL, Poewe W, Jankovic J. Parkinson disease and the immune system—Associations, mechanisms and therapeutics. Nat Rev Neurol. 2020;16:303–318. [DOI] [PubMed] [Google Scholar]
- 64. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16:626–638. [DOI] [PubMed] [Google Scholar]
- 65. Takahashi T, Iwasaki A. Sex differences in immune responses. Science. 2021;371:347–348. [DOI] [PubMed] [Google Scholar]
- 66. Kodama L, Gan L. Do microglial sex differences contribute to sex differences in neurodegenerative diseases? Trends Mol Med. 2019;25:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lynch MA. Exploring sex-related differences in microglia may be a game-changer in precision medicine. Front Aging Neurosci. 2022;14:868448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging. 2003;24:197–211. [DOI] [PubMed] [Google Scholar]
- 69. Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of Parkinson's disease. Lancet Neurol. 2020;19:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zach S, Felk S, Gillardon F. Signal transduction protein array analysis links LRRK2 to Ste20 kinases and PKC zeta that modulate neuronal plasticity. PLoS One. 2010;5:e13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gomez-Suaga P, Luzon-Toro B, Churamani D, et al. . Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum Mol Genet. 2012;21:511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Piccoli G, Onofri F, Cirnaru MD, et al. . Leucine-rich repeat kinase 2 binds to neuronal vesicles through protein interactions mediated by its C-terminal WD40 domain. Mol Cell Biol. 2014;34:2147–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen ZC, Zhang W, Chua LL, et al. . Phosphorylation of amyloid precursor protein by mutant LRRK2 promotes AICD activity and neurotoxicity in Parkinson's disease. Sci Signal. 2017;10:eaam6790. [DOI] [PubMed] [Google Scholar]
- 74. Meskini R E, Delfino C, Boudouresque F, Oliver C, Martin PM, Ouafik LH. Evidence of high expression of peptidylglycine alpha-amidating monooxygenase in the rat uterus: Estrogen regulation. Proc Natl Acad Sci U S A. 1998;95:7191–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stygelbout V, Leroy K, Pouillon V, et al. . Inositol trisphosphate 3-kinase B is increased in human Alzheimer brain and exacerbates mouse Alzheimer pathology. Brain. 2014;137(Pt 2):537–552. [DOI] [PubMed] [Google Scholar]
- 76. Paranjpe MD, Belonwu S, Wang JK, et al. . Sex-Specific cross tissue meta-analysis identifies immune dysregulation in women with Alzheimer's disease. Front Aging Neurosci. 2021;13:735611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Breuza L, Halbeisen R, Jeno P, et al. . Proteomics of Endoplasmic Reticulum-Golgi Intermediate Compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem. 2004;279:47242–47253. [DOI] [PubMed] [Google Scholar]
- 78. Colla E. Linking the endoplasmic Reticulum to Parkinson's disease and alpha-synucleinopathy. Front Neurosci. 2019;13:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Saito Y, Santosa V, Ishiguro KI, Kanemaki MT. MCMBP Promotes the assembly of the MCM2-7 hetero-hexamer to ensure robust DNA replication in human cells. Elife. 2022;11:e77393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kusters CD, Paul KC, Duarte Folle A, et al. . Increased menopausal age reduces the risk of Parkinson's disease: A Mendelian randomization approach. Mov Disord. 2021;36:2264–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Laws KR, Irvine K, Gale TM. Sex differences in Alzheimer's disease. Curr Opin Psychiatry. 2018;31:133–139. [DOI] [PubMed] [Google Scholar]
- 82. Vetvik KG, MacGregor EA. Sex differences in the epidemiology, clinical features, and pathophysiology of migraine. Lancet Neurol. 2017;16:76–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are openly available from an online repository or are available on request from study authors. RNA-sequencing data can be accessed through the Gene Expression Omnibus (accession ID: GSE216281). All code is freely available at https://github.com/chiaracapp/Gene-Expression-and-Genetic-Sex-Differences-in-PD.git Analyses were conducted in Python v.3.5, MATLAB R2020b and R v.4.2.2. Locus definition, functional annotation and gene-set analysis were performed using FUMA (https://fuma.ctglab.nl/).40