Fig. 5.
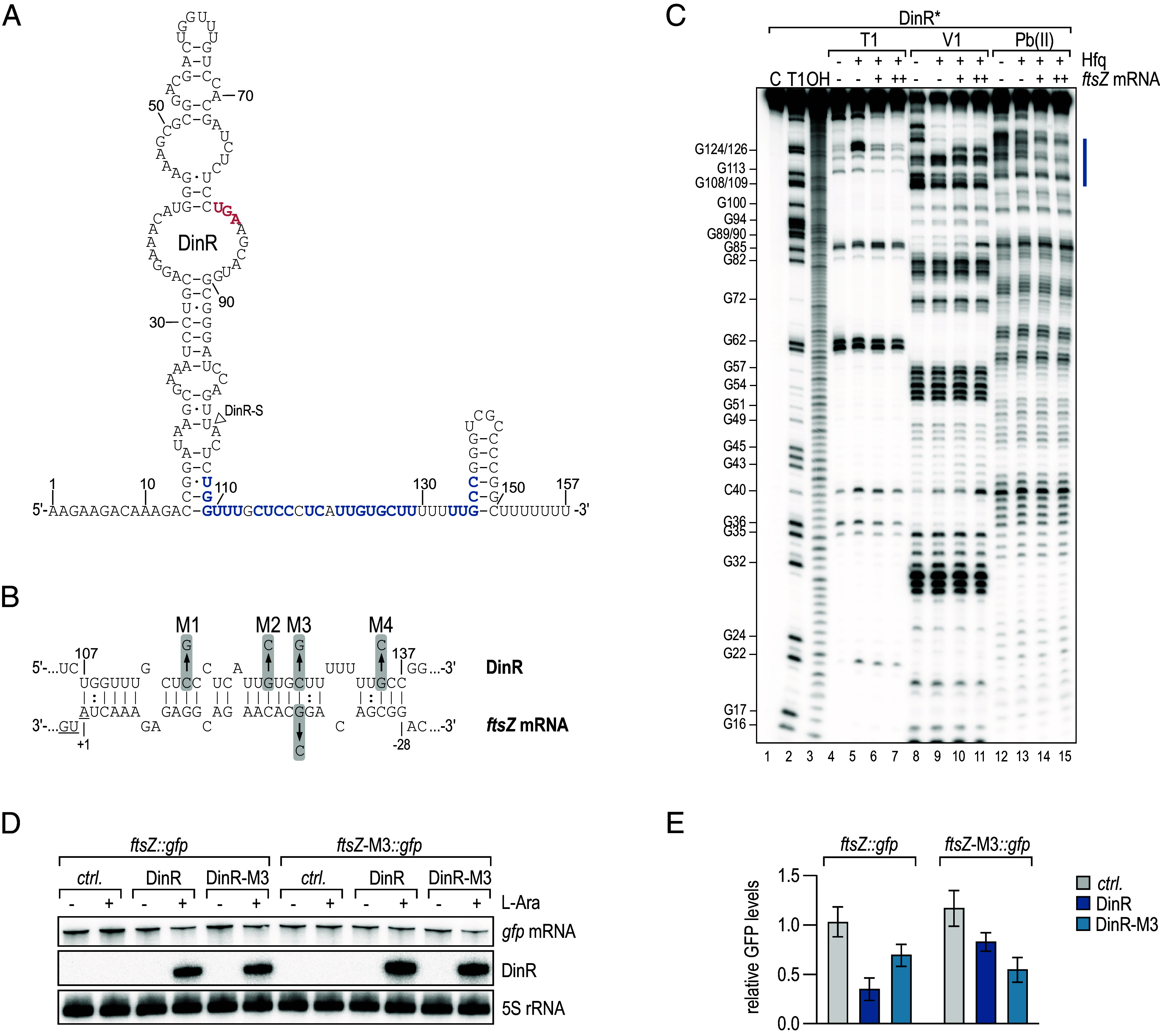
DinR represses ftsZ mRNA through a direct RNA–RNA interaction. (A) Secondary structure of DinR as determined by bioinformatics predictions and chemical probing shown in (C). The dinI stop codon is marked in red; the nucleotides predicted to interact with ftsZ mRNA are highlighted in blue. The 5′ end of DinR-S is indicated by a triangle. (B) Predicted base-pairing interaction forming between DinR and ftsZ mRNA. For DinR, the positions are numbered relative to the sRNA start site. For ftsZ mRNA, the positions are numbered relative to the start codon (underlined). Positions of single-nucleotide exchanges generating mutants M1, M2, M4, and the compensatory mutant M3 are indicated. (C) In vitro structure probing of 5′-end-labeled DinR (0.4 pmol) sRNA with RNase T1 (lanes 4 to 7), RNase V1 (lanes 8 to 11), and lead(II) acetate (lanes 12 to 15) in the absence (−) or presence (+) of 5x Hfq protein and 10x (+) or 25x (++) ftsZ mRNA. RNase T1 and alkaline ladders of DinR were used to map the positions of individual nucleotides. The putative ftsZ mRNA binding site is marked in blue. (D) E. coli carrying either an empty control vector pBADEC-ctrl., pBADEC-DinR or pBADEC-DinR-M3 in combination with the posttranscriptional reporters ftsZ::gfp or ftsZ-M3::gfp, respectively, were cultivated to OD600 of 0.5, and then, expression from the araBAD promoter was induced. RNA samples collected prior to and 5 min after addition of arabinose were analyzed by Northern blotting to determine the expression of DinR and ftsZ::gfp mRNA and their respective variants; 5S rRNA served as loading control. (E) Quantification of ftsZ::gfp or ftsZ-M3::gfp mRNA levels 5 min after addition of arabinose determined as described in (D). mRNA levels were determined relative to the expression levels prior to addition of the inducer; error bars represent the SD calculated from three independent biological replicates.
