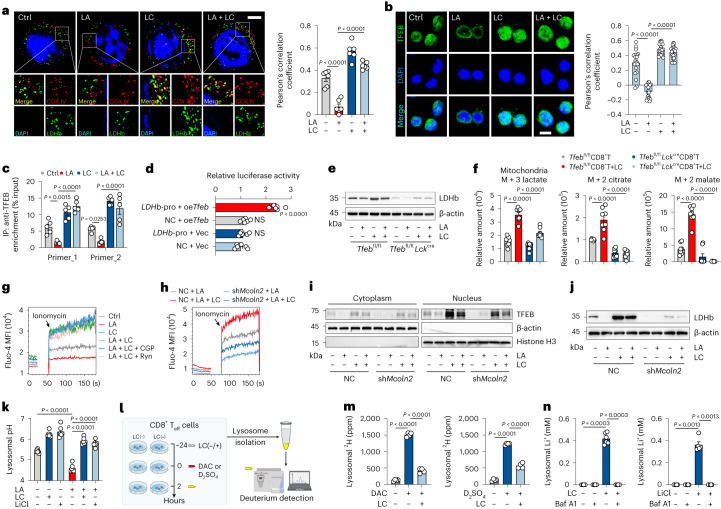
Fig. 3. LC triggers TFEB to transcriptionally activate LDHB.
a, Immunofluorescence staining of LDHB and COX IV in CD8+ Teff cells treated with LA and/or LC. Scale bar, 5 μm (left). Pearson’s correlation coefficient between LDHB and COX IV (right). n = 6 cells examined over three independent experiments. b, Immunofluorescence staining of TFEB in CD8+ Teff cells as in a. Scale bar, 10 μm (left). Pearson’s correlation coefficient between TFEB and DAPI (right). n = 20 cells examined over three independent experiments. c, ChIP–qPCR analysis of TFEB enrichment around the promoter of LDHB in CD8+ Teff cells. LDHB primer 1 and 2 were used. n = 5. d, Relative luciferase activity of HEK-293T cells transfected with Tfeb-overexpressing (oeTfeb) or LDHB-promoter (LDHB-pro) plasmids. n = 6. e, Immunoblots of LDHB in Tfebfl/fl or Tfebfl/fl Lckcre mice spleen-derived CD8+ Teff cells. f, Relative abundance of mitochondrial labeled lactate, citrate and malate derived from [13C]lactate in CD8+ Teff cells of Tfebfl/fl or Tfebfl/fl Lckcre mice. n = 7 in M + 2 citrate Tfebfl/fl CD8+ T group, n = 8 in other groups. g, Fluo-4 mean fluorescence intensity (MFI) of cytosolic calcium release in CD8+ Teff cells pretreated with CGP37157 (CGP, 10 μM) or ryanodine (Ryn, 100 μM) for 1 h then treated with LA and/or LC. h, Fluo-4 MFI of cytosolic calcium release in CD8+ Teff cells pre-transduced with shMcoln2 then treated with LA and/or LC. i,j, Immunoblots of TFEB (i) and LDHB (j) in CD8+ Teff cells as in h. k, Lysosomal pH value of CD8+ Teff cells was detected. n = 6. l, Schematic of experimental design for deuterium detection. Created with BioRender.com. m, Lysosomal 2H concentration, determined by isotope-MS, of CD8+ Teff cells with or without deuterated 2H-acetic acid (DAC, 10 mM) or 2H-sulfuric acid (D2SO4, 5 mM) for 3 h. n = 4. n, Lysosomal Li+ concentration in CD8+ Teff cells treated with LC/LiCl and/or bafilomycin A1 (Baf A1, 1 nM) was detected. n = 6 (left) and n = 5 (right). Data are the mean ± s.e.m. n = biological replicates unless stated otherwise. P values were calculated using one-way ANOVA for Dunnett’s multiple-comparisons test (a–d, f, k, and m) and Kruskal–Wallis test (n).